微生物组学的技术和方法及其应用
- 1中国科学院南京土壤研究所, 南京 210008
2中国科学院大学,北京 100049
收稿日期: 2019-08-19
录用日期: 2019-10-23
网络出版日期: 2020-02-24
基金资助
国家自然科学基金(31870480)
Techniques and methods of microbiomics and their applications
- 1Institute of Soil Science, Chinese Academy of Sciences, Nanjing 210008, China
2University of Chinese Academy of Sciences, Beijing 100049, China
Received date: 2019-08-19
Accepted date: 2019-10-23
Online published: 2020-02-24
Supported by
National Natural Science Foundation of China(31870480)
摘要
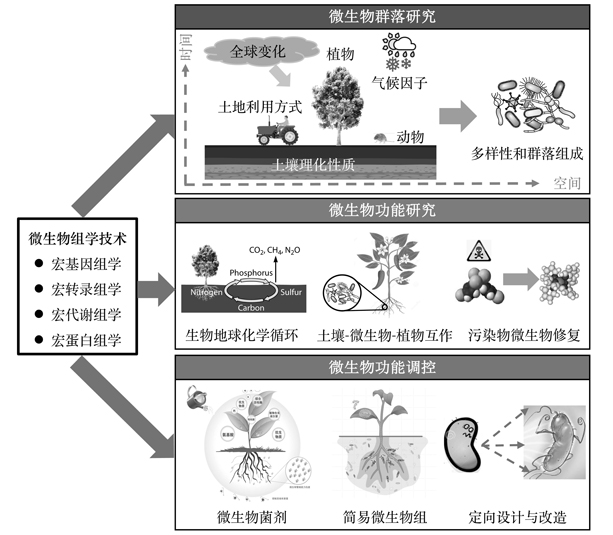
微生物组是指一个特定环境或生态系统中全部微生物及其遗传信息的集合, 其蕴藏着极为丰富的微生物资源。全面系统地解析微生物组的结构和功能, 将为解决人类面临的能源、生态环境、工农业生产和人体健康等重大问题带来新思路。然而, 微生物组学研究在很大程度上取决于其技术与方法的发展。在高通量测序技术出现以前, 微生物研究主要基于分离培养和指纹图谱等技术, 然而, 由于这些技术存在的缺陷, 人们对于微生物的认识十分有限。自21世纪初以来, 尽管高通量测序和质谱技术的革命性突破极大地促进了人们对于微生物的认识, 微生物组学技术在微生物组研究中的应用仍面临着诸多挑战。此外, 目前微生物组的结构和多样性等描述性研究已臻成熟, 微生物组学研究正处于从数量到质量、从结构到功能的关键转变时期。因此, 该文首先介绍了微生物组学的基本概念及其发展简史, 其次简述了微生物组学研究的相关技术和方法及其发展历程, 并进一步阐述了微生物组学的技术和方法在生态学研究中的应用及存在的主要问题, 最后从技术、理论和应用层面阐述了未来微生物组学技术和方法发展的前沿方向, 并提出了今后微生物组学研究的优先发展领域。
本文引用格式
高贵锋, 褚海燕 . 微生物组学的技术和方法及其应用[J]. 植物生态学报, 2020 , 44(4) : 395 -408 . DOI: 10.17521/cjpe.2019.0222
Abstract
Microbiome is the combination of all microorganisms and their genetic information in a specific environment or ecosystem, which contains abundant microbial resources. A comprehensive and systematic analysis of the structure and function of microbiome will provide new ideas in solving the core issues in the fields of energy, ecological environment, industrial and agricultural production and human health. However, the study of microbiome largely depends on the development of relevant technologies and methods. Before to the advent of high-throughput sequencing technology, microbial research was mainly based on techniques such as isolation, pure-culture and fingerprint. However, due to the technical restrictions, scientists could only get limited knowledge of microorganisms. Since the beginning of 21st century, the revolutionary advances in the technology of high-throughput sequencing and mass spectrometry have greatly improved our understanding on the structure and ecological functions of environmental microbiome. However, the application of microbiomics technology in microbial research still faces many challenges. In addition, the descriptive studies focusing on the structure and diversity of microbiome have already matured, and the study of microbiomics is facing a critical transition period from quantity to quality and from structure to function. Hence, this paper will firstly introduce the basic concepts of microbiomics and a brief development history. Secondly, this paper introduces the related technologies and methods of microbiomics with their development process, and further expounds the applications and main problems of microbiomics technologies and methods in ecological study. Finally, this paper expounds the frontier direction of the development of microbiomics technology and methods from the technical, theoretical and application levels, and proposes the priority development areas of microbiome research in the future.

参考文献
| [1] | Aguiar-Pulido V, Huang WR, Suarez-Ulloa V, Cickovski T, Mathee K, Narasimhan G (2016). Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis. Evolutionary Bioinformatics, 12(S1), 5-16. |
| [2] | Anesio AM, Lutz S, Chrismas NAM, Benning LG (2017). The microbiome of glaciers and ice sheets. NPJ Biofilms and Microbiomes, 3, 10. DOI: 10.1038/s41522-017-0019-0. |
| [3] | Antony-Babu S, Murat C, Deveau A, Le Tacon F, Frey-Klett P, Uroz S (2013). An improved method compatible with metagenomic analyses to extract genomic DNA from soils in Tuber melanosporum orchards. Journal of Applied Microbiology, 115, 163-170. |
| [4] | Bahram M, Hildebrand F, Forslund SK, Anderson JL, Soudzilovskaia NA, Bodegom PM, Bengtsson-Palme J, Anslan S, Coelho LP, Harend H, Huerta-Cepas J, Medema MH, Maltz MR, Mundra S, Olsson PA, Pent M, P?lme S, Sunagawa S, Ryberg M, Tedersoo L, Bork P (2018). Structure and function of the global topsoil microbiome. Nature, 560, 233-237. |
| [5] | Bailly J, Fraissinet-Tachet L, Verner MC, Debaud JC, Lemaire M, Wésolowski-Louvel M, Marmeisse R (2007). Soil eukaryotic functional diversity, a metatranscriptomic approach. The ISME Journal, 1, 632-642. |
| [6] | Bashiardes S, Zilberman-Schapira G, Elinav E (2016). Use of metatranscriptomics in microbiome research. Bioinformatics and Biology Insights, 10, 19-25. |
| [7] | Bastida F, Hernández T, García C (2014). Metaproteomics of soils from semiarid environment: functional and phylogenetic information obtained with different protein extraction methods. Journal of Proteomics, 101, 31-42. |
| [8] | Becher D, Bernhardt J, Fuchs S, Riedel K (2013). Metaproteomics to unravel major microbial players in leaf litter and soil environments: challenges and perspectives. Proteomics, 13, 2895-2909. |
| [9] | Benndorf D, Balcke GU, Harms H, von Bergen M (2007). Functional metaproteome analysis of protein extracts from contaminated soil and groundwater. The ISME Journal, 1, 224-234. |
| [10] | Breitwieser FP, Lu J, Salzberg SL (2019). A review of methods and databases for metagenomic classification and assembly. Briefings in Bioinformatics, 20, 1125-1136. |
| [11] | ?apek P, Manzoni S, Ka?tovská E, Wild B, Diáková K, Bárta J, Schnecker J, Biasi C, Martikainen PJ, Alves RJE, Guggenberger G, Gentsch N, Hugelius G, Palmtag J, Mikutta R, Shibistova O, Urich T, Schleper C, Richter A, ?antr??ková H (2018). A plantmicrobe interaction framework explaining nutrient effects on primary production. Nature Ecology & Evolution, 2, 1588-1596. |
| [12] | Damon C, Lehembre F, Oger-Desfeux C, Luis P, Ranger J, Fraissinet-Tachet L, Marmeisse R (2012). Metatranscriptomics reveals the diversity of genes expressed by eukaryotes in forest soils. PLOS ONE, 7, e28967. DOI: 10.1371/journal.pone.0028967. |
| [13] | Deng YL, Ruan YJ, Ma B, Timmons MB, Lu HF, Xu XY, Zhao HP, Yin XW (2019). Multiomics analysis reveals niche and fitness differences in typical denitrification microbial aggregations. Environment International, 132, 105085. DOI: 10.1016/j.envint.2019.105085. |
| [14] | Egert M, de Graaf AA, Smidt H, de Vos WM, Venema K (2006). Beyond diversity: functional microbiomics of the human colon. Trends in Microbiology, 14, 86-91. |
| [15] | Emerson JB, Adams RI, Román CMB, Brooks B, Coil DA, Dahlhausen K, Ganz HH, Hartmann EM, Hsu T, Justice NB, Paulino-Lima IG, Luongo JC, Lymperopoulou DS, Gomez-Silvan C, Rothschild-Mancinelli B, Balk M, Huttenhower C, Nocker A, Vaishampayan P, Rothschild LJ (2017). Schr?dinger’s microbes: tools for distinguishing the living from the dead in microbial ecosystems. Microbiome, 5, 86. DOI: 10.1186/s40168-017-0285-3. |
| [16] | Falkowski PG, Fenchel T, Delong EF (2008). The microbial engines that drive Earth’s biogeochemical cycles. Science, 320, 1034-1039. |
| [17] | Feng MM, Adams JM, Fan KK, Shi Y, Sun RB, Wang DZ, Guo XS, Chu HY (2018). Long-term fertilization influences community assembly processes of soil diazotrophs. Soil Biology & Biochemistry, 126, 151-158. |
| [18] | Gilbert JA, Jansson JK, Knight R (2018). Earth microbiome project and global systems biology. mSystems, 3, e00217-17. DOI: 10.1128/mSystems.00217-17. |
| [19] | Gilbert JA, Thomas S, Cooley NA, Kulakova A, Field D, Booth T, McGrath JW, Quinn JP, Joint I (2009). Potential for phosphonoacetate utilization by marine bacteria in temperate coastal waters. Environmental Microbiology, 11, 111-125. |
| [20] | Gohl DM, Vangay P, Garbe J, MacLean A, Hauge A, Becker A, Gould TJ, Clayton JB, Johnson TJ, Hunter R, Knights D, Beckman KB (2016). Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nature Biotechnology, 34, 942-949. |
| [21] | Gorka S, Dietrich M, Mayerhofer W, Gabriel R, Wiesenbauer J, Martin V, Zheng Q, Imai B, Prommer J, Weidinger M, Schweiger P, Eichorst SA, Wagner M, Richter A, Schintlmeister A, Woebken D, Kaiser C (2019). Rapid transfer of plant photosynthates to soil bacteria via ectomycorrhizal hyphae and its interaction with nitrogen availability. Frontiers in Microbiology, 10, 168. DOI: 10.3389/fmicb.2019.00168. |
| [22] | Guo X, Feng JJ, Shi Z, Zhou XS, Yuan MT, Tao XY, Hale L, Yuan T, Wang JJ, Qin YJ, Zhou AF, Fu Y, Wu LY, He ZL, van Nostrand JD, Ning DL, Liu XD, Luo YQ, Tiedje JM, Yang YF, Zhou JZ (2018). Climate warming leads to divergent succession of grassland microbial communities. Nature Climate Change, 8, 813-818. |
| [23] | Gutierrez T, Biddle JF, Teske A, Aitken MD (2015). Cultivation- dependent and cultivation-independent characterization of hydrocarbon-degrading bacteria in Guaymas Basin sediments. Frontiers in Microbiology, 6, 695. DOI: 10.3389/fmicb.2015.00695. |
| [24] | Han J, Antunes LCM, Finlay BB, Borchers CH (2010). Metabolomics: towards understanding host-microbe interactions. Future Microbiology, 5, 153-161. |
| [25] | Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM (1998). Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chemistry & Biology, 5, 245-249. |
| [26] | He JZ, Lu YH, Fu BJ (2014). Frontiers in Soil Biology. Science Press, Beijing. |
| [26] | [ 贺纪正, 陆雅海, 傅伯杰 (2014). 土壤生物学前沿. 科学出版社, 北京.] |
| [27] | Herbst FA, Taubert M, Jehmlich N, Behr T, Schmidt F, von Bergen M, Seifert J (2013). Sulfur-34S stable isotope labeling of amino acids for quantification (SULAQ34) of proteomic changes in Pseudomonas fluorescens during naphthalene degradation. Molecular & Cellular Proteomics, 12, 2060-2069. |
| [28] | Hill EM, Robinson LA, Abdul-Sada A, Vanbergen AJ, Hodge A, Hartley SE (2018). Arbuscular mycorrhizal fungi and plant chemical defence: effects of colonisation on aboveground and belowground metabolomes. Journal of Chemical Ecology, 44, 198-208. |
| [29] | Horner-Devine MC, Carney KM, Bohannan BJM (2004a). An ecological perspective on bacterial biodiversity. Proceedings of the Royal Society B: Biological Sciences, 271, 113-122. |
| [30] | Horner-Devine MC, Lage M, Hughes JB, Bohannan BJM (2004b). A taxa-area relationship for bacteria. Nature, 432, 750-753. |
| [31] | Huang WJ, Hammel KE, Hao JL, Thompson A, Timokhin VI, Hall SJ (2019). Enrichment of lignin-derived carbon in mineral-associated soil organic matter. Environmental Science & Technology, 53, 7522-7531. |
| [32] | Jansson JK, Baker ES (2016). A multi-omic future for microbiome studies. Nature Microbiology, 1, 16049. DOI: 10.1038/nmicrobiol.2016.49. |
| [33] | Jones MB, Highlander SK, Anderson EL, Li WZ, Dayrit M, Klitgord N, Fabani MM, Seguritan V, Green J, Pride DT, Yooseph S, Biggs W, Nelson KE, Venter JC (2015). Library preparation methodology can influence genomic and functional predictions in human microbiome research. Proceedings of the National Academy of Sciences of the United States of America, 112, 14024-14029. |
| [34] | Kleiner M, Dong XL, Hinzke T, Wippler J, Thorson E, Mayer B, Strous M (2018). Metaproteomics method to determine carbon sources and assimilation pathways of species in microbial communities. Proceedings of the National Academy of Sciences of the United States of America, 115, 5576-5584. |
| [35] | Kyrpides NC, Eloe-Fadrosh EA, Ivanova NN (2016). Microbiome data science: understanding our microbial planet. Trends in Microbiology, 24, 425-427. |
| [36] | Ladau J, Shi Y, Jing X, He JS, Chen LT, Lin XG, Fierer N, Gilbert JA, Pollard KS, Chu HY (2018). Existing climate change will lead to pronounced shifts in the diversity of soil prokaryotes. mSystems, 3, e00167-18. DOI: 10.1128/mSystems.00167-18. |
| [37] | Langille MGI, Ravel J, Fricke WF (2018). “Available upon request”: not good enough for microbiome data! Microbiome, 6, 8. DOI: 10.1186/s40168-017-0394-z. |
| [38] | Lawson CE, Harcombe WR, Hatzenpichler R, Lindemann SR, L?ffler FE, O’Malley MA, García Martín H, Pfleger BF, Raskin L, Venturelli OS, Weissbrodt DG, Noguera DR, McMahon KD (2019). Common principles and best practices for engineering microbiomes. Nature Reviews Microbiology, 17, 725-741. |
| [39] | Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature, 442, 806-809. |
| [40] | Li XG, Jousset A, de Boer W, Carrión VJ, Zhang TL, Wang XX, Kuramae EE (2019). Legacy of land use history determines reprogramming of plant physiology by soil microbiome. The ISME Journal, 13, 738-751. |
| [41] | Liu GH, Ye ZF, Wu WZ (2012). Culture-dependent and culture-independent approaches to studying soil microbial diversity. Acta Ecologica Sinica, 32, 4421-4433. |
| [41] | [ 刘国华, 叶正芳, 吴为中 (2012). 土壤微生物群落多样性解析法: 从培养到非培养. 生态学报, 32, 4421-4433.] |
| [42] | Liu SJ, Shi WY, Zhao GP (2017). China microbiome initiative: opportunity and challenges. Bulletin of Chinese Academy of Sciences, 32, 241-250. |
| [42] | [ 刘双江, 施文元, 赵国屏 (2017). 中国微生物组计划: 机遇与挑战. 中国科学院院刊, 32, 241-250.] |
| [43] | Liu YX, Qin Y, Guo XX, Bai Y (2019). Methods and applications for microbiome data analysis. Hereditas, 41, 845-862. |
| [43] | [ 刘永鑫, 秦媛, 郭晓璇, 白洋 (2019). 微生物组数据分析方法与应用. 遗传, 41, 845-862.] |
| [44] | Lupatini M, Suleiman AKA, Jacques RJS, Lemos LN, Pylro VS, van Veen JA, Kuramae EE, Roesch LFW (2019). Moisture is more important than temperature for assembly of both potentially active and whole prokaryotic communities in subtropical grassland. Microbial Ecology, 77, 460-470. |
| [45] | Ma JC, Zhao FQ, Su XQ, Xu J, Wu LH (2017). Strategies on establishment of China’s microbiome data center. Bulletin of Chinese Academy of Sciences, 32, 290-296. |
| [45] | [ 马俊才, 赵方庆, 苏晓泉, 徐健, 吴林寰 (2017). 关于中国微生物组数据中心建设的思考. 中国科学院院刊, 32, 290-296.] |
| [46] | Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, ?vre?s L, Reysenbach AL, Smith VH, Staley JT (2006). Microbial biogeography: putting microorganisms on the map. Nature Reviews Microbiology, 4, 102-112. |
| [47] | Moran MA (2015). The global ocean microbiome. Science, 350, aac8455. DOI: 10.1126/science.aac8455. |
| [48] | Moreno JL, Torres IF, García C, López-Mondéjar R, Bastida F (2019). Land use shapes the resistance of the soil microbial community and the C cycling response to drought in a semi-arid area. Science of the Total Environment, 648, 1018-1030. |
| [49] | Müller DB, Vogel C, Bai Y, Vorholt JA (2016). The plant microbiota: systems-level insights and perspectives. Annual Review of Genetics, 50, 211-234. |
| [50] | Musat N, Foster R, Vagner T, Adam B, Kuypers MMM (2012). Detecting metabolic activities in single cells, with emphasis on nano-SIMS. FEMS Microbiology Reviews, 36, 486-511. |
| [51] | Nesme J, Achouak W, Agathos SN, Bailey M, Baldrian P, Brunel D, Frosteg?rd ?, Heulin T, Jansson JK, Jurkevitch E, Kruus KL, Kowalchuk GA, Lagares A, Lappin-Scott HM, Lemanceau P, Le Paslier D, Mandic-Mulec I, Murrell JC, Myrold DD, Nalin R, Nannipieri P, Neufeld JD, O’Gara F, Parnell JJ, Pühler A, Pylro V, Ramos JL, Roesch LFW, Schloter M, Schleper C, Sczyrba A, Sessitsch A, Sj?ling S, S?rensen J, S?rensen SJ, Tebbe CC, Topp E, Tsiamis G, van Elsas JD, van Keulen G, Widmer F, Wagner M, Zhang T, Zhang XJ, Zhao LP, Zhu YG, Vogel TM, Simonet P (2016). Back to the future of soil metagenomics. Frontiers in Microbiology, 7, 73. DOI: 10.3389/fmicb.2016.00073. |
| [52] | O’ Malley MA (2007). The nineteenth century roots of “everything is everywhere”. Nature Reviews Microbiology, 5, 647-651. |
| [53] | Poretsky RS, Hewson I, Sun SL, Allen AE, Zehr JP, Moran MA (2009). Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environmental Microbiology, 11, 1358-1375. |
| [54] | Poussin C, Sierro N, Boué S, Battey J, Scotti E, Belcastro V, Peitsch MC, Ivanov NV, Hoeng J (2018). Interrogating the microbiome: experimental and computational considerations in support of study reproducibility. Drug Discovery Today, 23, 1644-1657. |
| [55] | Raut S, Polley HW, Fay PA, Kang S (2018). Bacterial community response to a preindustrial-to-future CO2 gradient is limited and soil specific in Texas Prairie grassland. Global Change Biology, 24, 5815-5827. |
| [56] | Rice MC, Norton JM, Valois F, Bollmann A, Bottomley PJ, Klotz MG, Laanbroek HJ, Suwa Y, Stein LY, Sayavedra-Soto L, Woyke T, Shapiro N, Goodwin LA, Huntemann M, Clum A, Pillay M, Kyrpides N, Varghese N, Mikhailova N, Markowitz V, Palaniappan K, Ivanova N, Stamatis D, Reddy TBK, Ngan CY, Daum C (2016). Complete genome of Nitrosospira briensis C-128, an ammonia-oxidizing bacterium from agricultural soil. Standards in Genomic Sciences, 11, 46. DOI: 10.1186/s40793-016-0168-4. |
| [57] | Rime T, Hartmann M, Brunner I, Widmer F, Zeyer J, Frey B (2015). Vertical distribution of the soil microbiota along a successional gradient in a glacier forefield. Molecular Ecology, 24, 1091-1108. |
| [58] | Rodrigues JLM, Pellizari VH, Mueller R, Baek K, Jesus EDC, Paula FS, Mirza B, Hamaoui GS, Tsai SM, Feigl B, Tiedje JM, Bohannan BJM, Nüsslein K (2013). Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proceedings of the National Academy of Sciences of the United States of America, 110, 988-993. |
| [59] | Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M (1977). Nucleotide sequence of bacteriophage phi X174 DNA. Nature, 265, 687-695. |
| [60] | Schadt EE, Turner S, Kasarskis A (2010). A window into third-generation sequencing. Human Molecular Genetics, 19, 227-240. |
| [61] | Schloss PD (2018). Identifying and overcoming threats to reproducibility, replicability, robustness, and generalizability in microbiome research. mBio, 9, e00525-18. DOI: 10.1128/mBio.00525-18. |
| [62] | Schloss PD, Jenior ML, Koumpouras CC, Westcott SL, Highlander SK (2016). Sequencing 16S rRNA gene fragments using the PacBio SMRT DNA sequencing system. PeerJ, 4, e1869. DOI 10.7717/peerj.1869. |
| [63] | Shendure J, Ji H (2008). Next-generation DNA sequencing. Nature Biotechnology, 26, 1135-1145. |
| [64] | Shi WC, Li MC, Wei GS, Tian RM, Li CP, Wang B, Lin RS, Shi CY, Chi XL, Zhou B, Gao Z (2019a). The occurrence of potato common scab correlates with the community composition and function of the geocaulosphere soil microbiome. Microbiome, 7, 14. DOI: 10.1186/s40168-019-0629-2. |
| [65] | Shi WY, Qi HY, Sun QL, Fan GM, Liu SJ, Wang J, Zhu BL, Liu HW, Zhao FQ, Wang XC, Hu XX, Li W, Liu J, Tian Y, Wu LH, Ma JC (2019b). gcMeta: a global catalogue of metagenomics platform to support the archiving, standardization and analysis of microbiome data. Nucleic Acids Research, 47, 637-648. |
| [66] | Shi YM, Tyson GW, DeLong EF (2009). Metatranscriptomics reveals unique microbial small RNAs in the ocean?s water column. Nature, 459, 266-269. |
| [67] | Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ (2006). Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proceedings of the National Academy of Sciences of the United States of America, 103, 12115-12120. |
| [68] | Tang J (2011). Microbial metabolomics. Current Genomics, 12, 391-403. |
| [69] | Tedersoo L, Bahram M, P?lme S, K?ljalg U, Yorou NS, Wijesundera R, Ruiz LV, Vasco-Palacios AM, Thu PQ, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Rosas M, Riit T, Ratkowsky D, Pritsch K, P?ldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, P?rtel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Majuakim L, Lodge DJ, Lee SS, Larsson KH, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo LD, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, De Kesel A, Dang T, Chen X, Buegger F, Brearley FQ, Bonito G, Anslan S, Abell S, Abarenkov K (2014). Global diversity and geography of soil fungi. Science, 346, 1256688. DOI: 10.1126/science.1256688. |
| [70] | Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, Navas-Molina JA, Janssen S, Kopylova E, Vázquez- Baeza Y, González A, Morton JT, Mirarab S, Xu ZJ, Jiang LJ, Haroon MF, Kanbar J, Zhu QY, Jin Song S, Kosciolek T, Bokulich NA, Lefler J, Brislawn CJ, Humphrey G, Owens SM, Hampton-Marcell J, Berg-Lyons D, McKenzie V, Fierer N, Fuhrman JA, Clauset A, Stevens RL, Shade A, Pollard KS, Goodwin KD, Jansson JK, Gilbert JA, Knight R , The Earth Microbiome Project Consortium (2017). A communal catalogue reveals Earth’s multiscale microbial diversity. Nature, 551, 457-463. |
| [71] | Tkacz A, Hortala M, Poole PS (2018). Absolute quantitation of microbiota abundance in environmental samples. Microbiome, 6, 110. DOI: 10.1186/s40168-018-0491-7. |
| [72] | Tourna M, Stieglmeier M, Spang A, K?nneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proceedings of the National Academy of Sciences of the United States of America, 108, 8420-8425. |
| [73] | Tripathi BM, Stegen JC, Kim M, Dong K, Adams JM, Lee YK (2018). Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. The ISME Journal, 12, 1072-1083. |
| [74] | Turnbaugh PJ, Gordon JI (2008). An invitation to the marriage of metagenomics and metabolomics. Cell, 134, 708-713. |
| [75] | Vilanova C, Porcar M (2016). Are multi-omics enough? Nature Microbiology, 1, 16101. DOI: 10.1038/nmicrobiol.2016.101. |
| [76] | Wagner MR, Lundberg DS, del Rio TG, Tringe SG, Dangl JL, Mitchell-Olds T (2016). Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nature Communications, 7, 12151. DOI: 10.1038/ncomms12151. |
| [77] | Wang DZ, Kong LF, Li YY, Xie ZX (2016). Environmental microbial community proteomics: status, challenges and perspectives. International Journal of Molecular Sciences, 17, 1275. DOI: 10.3390/ijms17081275. |
| [78] | Wang GY, Zang B, Gu Z (2009). Development and application of the mass spectrometry. Modern Scientific Instruments, 6, 124-128. |
| [78] | [ 王桂友, 臧斌, 顾昭 (2009). 质谱仪技术发展与应用. 现代科学仪器, 6, 124-128.] |
| [79] | Wang L, Tian L (2019). Application of metaomics in wastewater treatment. Microbiology China, 46, 2370-2377. |
| [79] | [ 王琳, 田璐 (2019). 宏组学方法在污水处理系统中的应用进展. 微生物学通报, 46, 2370-2377.] |
| [80] | White III RA, Callister SJ, Moore RJ, Baker ES, Jansson JK (2016). The past, present and future of microbiome analyses. Nature Protocols, 11, 2049-2053. |
| [81] | Wu QL, Jiang HL (2017). China lake microbiome project. Bulletin of Chinese Academy of Sciences, 32, 273-279. |
| [81] | [ 吴庆龙, 江和龙 (2017). 中国湖泊微生物组研究. 中国科学院院刊, 32, 273-279.] |
| [82] | Xiang XJ, He D, He JS, Myrold DD, Chu HY (2017). Ammonia- oxidizing bacteria rather than archaea respond to short-term urea amendment in an alpine grassland. Soil Biology & Biochemistry, 107, 218-225. |
| [83] | Xu J, Ma B, Su XQ, Huang S, Xu X, Zhou XD, Huang WE, Knight R (2017). Emerging trends for microbiome analysis: from single-cell functional imaging to microbiome big data. Engineering, 3, 66-70. |
| [84] | Yang T, Adams JM, Shi Y, He JS, Jing X, Chen LT, Tedersoo L, Chu HY (2017). Soil fungal diversity in natural grasslands of the Tibetan Plateau: associations with plant diversity and productivity. New Phytologist, 215, 756-765. |
| [85] | Yang T, Tedersoo L, Soltis PS, Soltis DE, Gilbert JA, Sun M, Shi Y, Wang HF, Li YT, Zhang J, Chen ZD, Lin HY, Zhao YP, Fu CX, Chu HY (2019). Phylogenetic imprint of woody plants on the soil mycobiome in natural mountain forests of eastern China. The ISME Journal, 13, 686-697. |
| [86] | Yang YF (2013). Omics breakthroughs for environmental microbiology. Microbiology China, 40, 18-33. |
| [86] | [ 杨云锋 (2013). 环境微生物学的组学技术应用和突破. 微生物通报, 40, 18-33.] |
| [87] | Yu RT, Gao PJ, Han L, Huang LY (2009). Strategy and application of metaproteomics. Chinese Journal of Biotechnology, 25, 961-967. |
| [87] | [ 于仁涛, 高培基, 韩黎, 黄留玉 (2009). 宏蛋白质组学研究策略及应用. 生物工程学报, 25, 961-967.] |
| [88] | Zhang KP, Shi Y, Cui XQ, Yue P, Li KH, Liu XJ, Tripathi BM, Chu HY (2019). Salinity is a key determinant for soil microbial communities in a desert ecosystem. mSystems, 4, e00225-18. DOI: 10.1128.mSystems.00225-18. |
| [89] | Zhang KP, Shi Y, Jing X, He JS, Sun RB, Yang YF, Shade A, Chu HY (2016). Effects of short-term warming and altered precipitation on soil microbial communities in alpine grassland of the Tibetan Plateau. Frontiers in Microbiology, 7, 1032. DOI: 10.3389/fmicb.2016.01032. |
| [90] | Zhao Y, Gu RS, Du SM (2012). The research status and development tendency of bioinformatics. Journal of Medical Informatics, 33, 2-6. |
| [90] | [ 赵屹, 谷瑞升, 杜生明 (2012). 生物信息学研究现状及发展趋势. 医学信息学杂志, 33, 2-6.] |
| [91] | Zhong JN, Luo LJ, Chen BW, Sha S, Qing Q, Tam NFY, Zhang Y, Luan TG (2017). Degradation pathways of 1-methylphenanthrene in bacterial Sphingobium sp. MP9-4 isolated from petroleum-contaminated soil. Marine Pollution Bulletin, 114, 926-933. |
| [92] | Zhu YG, Shen RF, He JZ, Wang YF, Han XG, Jia ZJ (2017). China soil microbiome initiative: progress and perspective. Bulletin of Chinese Academy of Sciences, 32, 554-565. |
| [92] | [ 朱永官, 沈仁芳, 贺纪正, 王艳芬, 韩兴国, 贾仲君 (2017). 中国土壤微生物组: 进展与展望. 中国科学院院刊, 32, 554-565.] |