空气中约含80%的氮气, 因其化学惰性而无法被高等生物直接利用。只有固氮微生物具有将氮气转化为能被植物同化的氨的能力, 此即为生物固氮。据联合国粮农组织(FAO) 1995年粗略估计, 全球每年由生物固定的氮量已近2 × 1011 kg (相当于4 × 1011 kg尿素), 约占全球植物需氮量的3/4。所以, 生物固氮是地球上最大规模的天然氮肥加工厂。
根据固氮微生物与其他生物的关系, 可将生物固氮系统分为自生固氮、联合固氮和共生固氮3种不同的体系。其中, 共生固氮体系是自然界中主要的固氮体系, 包括根瘤菌与豆科植物、弗兰克氏菌与非豆科植物、固氮蓝细菌与植物(如满江红(Azolla imbricata)、苏铁(Cycas revoluta)和根乃拉草(Gunnera tinctoria)), 以及蓝细菌与真菌的地衣共生体等, 这一体系具有根瘤等特殊的共生结构, 固氮效率极高, 因而始终是生物固氮研究的焦点之一, 其中尤以豆科植物的共生固氮最为突出。世界上有豆科植物约19 700种, 其中已知可以结瘤固氮的有2 800多种, 占15% (Dénarié & Roche, 1991)。截止2003年, 全球种植豆科植物的面积达1.8亿hm2, 占地球可耕地的12%和世界主要农作物产量的27% (Graham & Vance, 2003)。它们非凡的共生固氮能力已使其成为人类食物蛋白的一个重要来源, 同时也是自然界和农业生态系统的重要氮素来源之一, 它们的次生代谢产物的重要价值也日益受到关注。此外, 豆科植物还能与真菌共生形成内生菌根。
豆科植物与根瘤菌共生互作的结果导致了一个新的植物器官——根瘤的形成。根瘤菌生活在根瘤中, 它们具有将氮气转化为能被植物同化的氨的能力。近年来, 人们对共生固氮的研究取得了长足的进展, 特别是植物方面涉及共生结瘤的研究已经全面展开, 主要表现在: 结瘤早期重要的信号传导过程初露端倪; 许多结瘤素基因从不同豆科植物中被分离筛选, 部分基因的功能得以阐明; 围绕模式植物蒺藜苜蓿(Medicago truncatula)和日本百脉根(Lotus japonicus)进行的结构、比较、功能基因组学及蛋白质组学的研究正如火如荼地展开。本文主要以蒺藜苜蓿和日本百脉根的各种突变株为例, 对近年来豆科植物在共生固氮方面特别是共生结瘤过程中的信号传导研究进展进行了综述, 并对当前存在的问题和以后的研究方向作了分析与展望, 以期为豆科植物与根瘤菌的共生互作研究提供参考。
1 根瘤的形成与类型
豆科植物可以与土壤中的根瘤菌属(Rhizo- bium)、慢生根瘤菌属(Bradyrhizobium)、中华根瘤菌属(Sinorhizobium)、中慢生根瘤菌属(Mesorhi- zobium)及固氮根瘤菌属(Azorhizobium)等根瘤菌共生结瘤, 这些共生的革兰氏阴性菌伙伴被统称为根瘤菌, 它们之间的共生互作起始于共生双方的分子对话。首先, 植物分泌类黄酮至根际, 诱导根瘤菌结瘤基因的表达并合成结瘤因子(Nod factor, NF)。这些结瘤因子是一些脂壳寡糖(lipochitooligosaccharides, LCOs), 它们被植物特异性地识别后, 植物的根迅速作出一系列反应, 如钙峰及离子流(Ca2+、Cl-、H+)的形成、根毛顶端膨胀和新的顶端生长、根毛分枝和卷曲(Dénarié et al., 1996; Cárdenas et al., 2000; Shaw & Long, 2003)。在卷曲的根毛内, 附着的根瘤菌进入细胞壁之间并开始形成侵入线。侵入线是一个管状结构, 由修饰过的植物细胞壁的局部降解及质膜内陷而成。与此同时, 某些皮层细胞进行有丝分裂, 根瘤原基开始形成(Timmers et al., 1999)。以类似顶端生长的方式, 侵入线在根毛内向前推进并穿过皮层细胞, 最后在根瘤原基细胞中释放根瘤菌, 后者进一步分化成被膜包围着的可以固氮的类菌体, 共生体形成, 根瘤原基逐渐发育为根瘤(van Brussel et al., 1992; van Spronsen et al., 1994; Brewin, 1998)。
结瘤因子主要由3-5个β-1, 4糖苷键连接的乙酰氨基葡萄糖寡聚体骨架和非还原端脂肪酸链组成, 其侧链上羟基的取代基团、脂酰链的长度及不饱和度决定着NF的活性和特异性, 并与根瘤菌的宿主专一性紧密相关。有效的共生侵入需要特殊取代基团的存在, 但这些取代基团并非对NF诱导的所有反应都是必需的。例如, 在蒺藜苜蓿与中华根瘤菌的共生互作中, 侵入线的形成严格要求结瘤因子葡糖胺骨架非还原端具有乙酰基, 但钙峰形成、一些特异基因的转录及根瘤原基的发生则没有这样严格的要求(Ardourel et al., 1994; Oldroyd et al., 2001; Wais et al., 2002)。因此, 有人提出了在Medicago中NF存在两类受体: 信号受体与进入受体, 前者是诱导结瘤过程早期反应所必需的, 后者则介导着根瘤菌在根表皮中的侵入过程, 对NF的结构有着更严格的要求(Ardourel et al., 1994)。
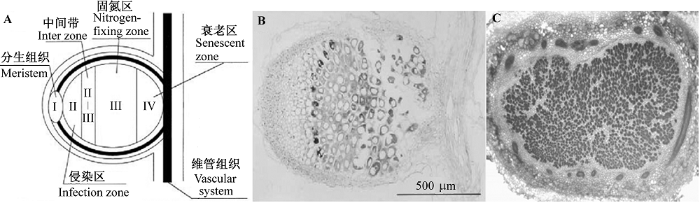
从结构上看, 根瘤一般可分为定型根瘤和不定型根瘤(图1)。前者主要在热带豆科植物如大豆(Glycine max)、豇豆(Vigna unguiculata)等的根上形成, 这类根瘤只在发育早期出现细胞分裂, 随后只进行细胞的伸长生长。在成熟的根瘤内, 包含固氮细胞的中央组织是均一的。不定型根瘤则通常形成于温带豆科植物上, 如蒺藜苜蓿、豌豆(Pisum sativum)、蚕豆(Vicia faba)及白花三叶草(Trifolium repens)等, 它们具有明显的分层: I层为顶端分生组织; II层为含有根瘤菌的侵染区; 然后是富含淀粉粒的II-III中间带, 主要的转录变化即发生于此; III层为固氮区, 其中充满能固氮的类菌体; 最后是接近衰老的IV层(Vasse et al., 1990; Brewin, 1991; Hirsch, 1992)。要注意的是, 日本百脉根虽然和蒺藜苜蓿同属于温带豆科植物, 但却形成定型根瘤。
图1
图1
定型根瘤与不定型根瘤示意图。
A, 不定型根瘤结构模式图。B, 紫云英不定型根瘤纵切图。C, 大豆定型根瘤纵切图。A和B来自Naito等(2000), C由华中农业大学农业微生物学国家重点实验室生物固氮研究室提供。
Fig. 1
Diagram of the determinate- and indeterminate-type nodule.
A, Schematic representation of the five distinct regions of an indeterminate type nodule. B, The vertical dissection of an indeterminate type nodule of Astragalus sinicus. C, The vertical dissection of a determinate type nodule of soybean. A and B are derived from Naito et al., 2000; C is provided by the laboratory of Biological Nitrogen Fixation, State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, China.
实际上, 定型根瘤与不定型根瘤的区别不仅表现在结构上, 在发育早期, 它们就已经显示了不同: 不定型根瘤的根瘤原基形成于内皮层, 而定型根瘤的根瘤原基形成于外皮层; 不定型根瘤早期伴随有胞质桥的发生, 而定型根瘤则不一定, 如菜豆(Phaseolus vulgaris)就不像日本百脉根一样形成胞质桥(van Spronsen et al., 2001)。另外, 类菌体在不同类型根瘤中的形成与代谢也各异。在定型根瘤中, 共生体间可以相互融合, 或者同一共生体中的类菌体进一步分裂, 结果形成几个类菌体共存于一个共生体中的局面; 而在不定型根瘤中, 单个的共生体连同类菌体一起分裂, 结果更多地是一个共生体中只包含一个类菌体(Prell & Poole, 2006)。研究表明, 在侵入线发育过程中, 聚羟基丁酸(PHB)在根瘤菌细胞中大量贮存(Lodwig et al., 2003, 2005)。在不定型根瘤的类菌体形成过程中, PHB颗粒裂解; 同时, 以不产PHB的根瘤菌突变体接种豌豆, 其不定型根瘤中的类菌体在发育过程中消耗了更多的植物淀粉(Lodwig et al., 2005), 表明PHB在类菌体形成中作为碳源和能源被利用, 分解代谢的急剧增强导致了PHB的消耗。但在定型根瘤中, 即使是成熟的类菌体, 仍然继续积累着大量的PHB。类菌体固定形成的NH4+进入植物细胞后, 在许多豆科植物特别是结不定型根瘤的植物细胞中, NH4+被结合形成谷氨酰胺或天冬酰胺, 而在热带结定型根瘤的豆科植物中, NH4+被结合为酰脲(Lodwig & Poole, 2003)。
然而, 无论哪种根瘤, 其形成和发育都离不开植物基因的调控和参与。van Kammen (1984)将在结瘤过程中被诱导表达或表达增强了的植物基因定义为结瘤素基因, 表达产物称为结瘤素。根据表达时间的早晚, 结瘤素基因可以分为早期结瘤素基因和晚期结瘤素基因, 前者在开始固氮前表达, 主要参与根瘤的形成和发育; 后者则在发育完全的、成熟的根瘤中表达, 与根瘤的功能有关, 包括一些参与初级代谢的酶、维持根瘤完整结构的蛋白质等, 其中研究得较为清楚的是豆血红蛋白基因(Nap & Bisseling, 1990)。
因为植株个体矮小、生育期短、基因组小、自花授粉和可被转化等优点, 蒺藜苜蓿和日本百脉根分别作为不定型根瘤和定型根瘤结瘤系统的代表植物而受到广泛研究。利用反向遗传学手段, 人们从这两种模式植物中获得了各种结瘤缺失突变植株, 从而开始了结瘤信号识别途径的研究。这些突变株一般分为两类: 不能结瘤的突变株Nod-和不能固氮的突变株Fix-。此外, 还有超结瘤突变株。
2 根瘤形成早期的信号传导
根瘤形成早期信号传导基因的分离主要通过对突变株的研究获得, 该类突变株不能形成根瘤、在结瘤因子信号传导中存在缺陷。最重要的突变之一应是来自日本百脉根的nfr1和nfr5突变株(Radutoiu et al., 2003), NFR1和NFR5基因编码LysM类丝氨酸/苏氨酸受体激酶(LysM RLKs)。LysM是一个肽聚糖结合结构域, 最初在细菌溶素和溶菌酶中发现, 一般存在于和细菌细胞壁及病原相关的蛋白质中。由于结瘤因子的基本结构是N-乙酰葡糖胺以β-1,4糖苷键相连的多聚体, 类似于肽聚糖的结构(N-乙酰胞壁酸和N-乙酰葡糖胺通过β-1,4糖苷键相连的多聚体), 因此, 虽然缺乏生化上的证据, NFR1和NFR5被公认为是直接与结瘤因子相作用的受体, 存在于质膜上。通过序列比较, 类似的突变株nfp (Nod factor perception)和sym10 (symbiosis 10), 相继在蒺藜苜蓿(Amor et al., 2003)和豌豆(Walker et al., 2000)中发现, 其中的相关基因同样编码LysM RLKs。在LysM类受体激酶突变株中, 所有结瘤因子诱导的反应(包括钙流和钙峰这一结瘤因子信号传导途径中最早出现的反应)均被破坏。
在蒺藜苜蓿中, 还发现一组不能形成侵染(do not make infections)的突变株: dmi1、dmi2、dmi3, dmi1和dmi2突变株不能被结瘤因子诱导产生钙峰, 钙流的存在也较野生型短暂(Wais et al., 2000; Shaw & Long, 2003; Ané et al., 2004)。DMI1由氨基端序列介导定位于蒺藜苜蓿根及根瘤细胞的核膜上, 羧基端包含一个钾离子电导率调控蛋白结构域(RCK), 与产甲烷杆菌中的钾离子通道蛋白(MthK)有着高度的相似性, 包括RCK在内的羧基端缺失突变明显干扰了根瘤的形成, 并使得G蛋白拮抗物胡蜂蜂毒素诱导的钙峰不能形成。研究者们认为, DMI1并没有直接参与结瘤因子诱导的钙离子变化, 而是调控着钙离子通道蛋白(Riely et al., 2007; Peiter et al., 2007)。DMI1在日本百脉根中有两个同源蛋白质: POLLUX和CASTOR, 它们的突变株虽能发生根毛分枝与变形, 但不发生根毛卷曲, 也不能形成侵入线; 与DMI1不同, POLLUX和CASTOR定位于质体上, Imaizumi-Anraku等(2005)推测它们可能形成复合物, 调节钙离子流穿过质体膜。近年的研究表明, POLLUX和CASTOR也为另一更古老的共生体系――植物与真菌的共生互作所需, 在豆科植物和非豆科植物中广泛存在且高度保守(Banba et al., 2008; Chen et al., 2009)。DMI2编码一个含有重复的亮氨酸丰富结构域的类受体激酶(LRR-RLK), 定位于质膜和侵入线膜上(Endre et al., 2002; Limpens et al., 2005), 它的同源物有: 来自日本百脉根和田菁(Sesbania cannabina)的SYMRK (symbiosis receptor kinase; Stracke et al., 2002; Capoen et al., 2005)、紫花苜蓿(Medicago sativa)的NORK (nodulation receptor kinase; Endre et al., 2002)和豌豆的SYM19 (Stracke et al., 2002)。dmi3编码钙离子/钙调素依赖的蛋白激酶(CCaMK) (Lévy et al., 2004), 日本百脉根中与其同源的基因为SNF1 (spontaneous nodule formation 1)。CCaMK含有一个自抑制结构域, 在与钙调素结合后自抑制结构域释放, 使得CCaMK的激酶活性得以表达, 从而激活根瘤的形态发生过程。和dmi1、dmi2一样, dmi3不能共生结瘤, 但该突变株能形成钙峰。在不接种根瘤菌的情况下, 自激活的CCaMK能诱导早期结瘤素基因ENOD11的表达, 并在植株上形成根瘤(Gleason et al., 2006; Tirichine et al., 2006)。除了负责转换钙峰信号以外, DMI3还可能通过磷酸化的方式控制NSP1的表达(Smit et al., 2005)。
nsp1、nsp2 (nodulation signaling pathway 2)具有和dmi3类似的突变表型, 遗传学分析表明, 它们位于dmi3的下游, 编码属于GRAS家族的蛋白质(Kaló et al., 2005; Smit et al., 2005)。GRAS结构域包含两个亮氨酸丰富区和一些小的稳定的保守序列, 如VHIID、PFYRE和SAW, 这个家族的成员与植物赤霉素和光敏色素的信号传导有关, 推测是一些转录调控因子。NSP1、DMI3均定位于根表皮细胞和皮层细胞的细胞核上(Smit et al., 2005), NSP2则定位在核膜和内质网上(Kaló et al., 2005)。研究表明, 蒺藜苜蓿经结瘤因子处理后, 一些早期结瘤素基因迅速表达, 其中包括ENOD11。自激活的CCaMK也能诱导ENOD11的表达, 同时还需要NSP1、NSP2及ERN1等基因参与调控(Gleason et al., 2006; Tirichine et al., 2006; Middleton et al., 2007), 表明CCaMK传递的钙信号正是通过上述NSP1、NSP2、ERN (ERF required for nodulation)等转录调控因子激活了靶基因的表达。ERN1及其同源蛋白ERN2、ERN3直接作用于ENOD11的启动子, 其中, ERN1和ERN2为转录激活物, 而ERN3则起抑制作用(Andriankaja et al., 2007)。最近的研究证实, 在体外, NSP1可直接结合于ENOD11、ERN1、NIN等早期结瘤素基因的启动子部位, 表明这些基因的诱导需要NSP1; 而在体内, NSP1与ENOD11启动子的相互作用还需要NSP2, 亦即以NSP1-NSP2异源多聚体的形式与之结合(Hirsch et al., 2009)。ERN类调控因子和NSP1与ENOD11启动子之间的结合位点仅相距26 bp, 推测这两类蛋白质之间可能存在着一定的相互作用。可见, 在NF信号传导途径中, 钙离子作为第二信使, 负责着宿主细胞质膜对根瘤菌NF的识别到细胞核中基因的表达变化之间的信号链接。
与ERN一样, EFD (ERF required for nodule differentiation)也属于乙烯响应因子群V (ethylene response factor group V, ERF V)的转录因子, 但是, EFD的氨基酸序列及表达方式均明显有别于ERN蛋白质(Vernié et al., 2008)。研究表明, EFD负调控着根瘤菌引起的侵染结瘤, 并为有固氮功能的根瘤形成所必需; 另外, EFD激活了细胞分裂素的初级受体基因Mt RR4的表达, 可能因而阻遏了细胞分裂素信号传递过程, 结果抑制了新的根瘤起始, 却促进了已起始根瘤分生组织细胞的分化(Vernié et al., 2008)。因此, EFD很可能是连接根瘤形成早期和晚期阶段的一个关键调控因子。来自日本百脉根的调控因子LjERF1属于另一个不同的乙烯响应因子群IX (Asamizu et al., 2008), LjERF1在根瘤菌Mesorhizobium loti对宿主的成功侵染过程中发挥着重要作用。接种后, LjERF1只在根的表皮细胞中表达, 在根瘤中没有任何表达, 推测是根瘤形成早期的正调控因子。
Kanamori等(2006)报道了一个核孔蛋白NUP133, 它的基因突变导致了日本百脉根的温敏型结瘤缺失, 即使在适宜的温度下, 也只能形成无效瘤, 侵入线中的少量根瘤菌不能被释放; 而且, 它对钙峰的诱导是必需的; NUP133定位于核膜上。因此, 在宿主植物识别相应的根瘤菌后, NUP133可能在核-质信息交流中起着一定的作用。在对上述突变株的研究中还发现, dmi类和nup133突变株也不能与菌根真菌共生, 真菌不能在植株体内定殖, 表明在这个古老的共生体系中, DMI及NUP类基因在菌根真菌的早期信号传导中同样起着重要作用(Endre et al., 2002; Stracke et al., 2002; Ané et al., 2004; Mitra et al., 2004; Lévy et al., 2004; Kanamori et al., 2006)。
其他重要的不结瘤突变株还有在根瘤发育或起始信号转换上存在缺陷的日本百脉根突变株nin (Schauser et al., 1999)、来自蒺藜苜蓿的不能发生根毛卷曲的hcl (Wais et al., 2000)等。这两种突变株虽然能够感知结瘤信号并表现根毛变形及显示钙峰, nin (nodule inception)甚至可以发生个别根毛的过度卷曲, 但它们不能形成侵入线、根瘤原基等。NIN含有跨膜结构域和一个DNA结合结构域, 推测是一个转录因子, 从豌豆中分离到它的同源基因SYM35 (Schauser et al., 1999; Borisov et al., 2003)。对蒺藜苜蓿同源突变株Mtnin的研究显示, NF早期信号传递过程并不需要NIN, 但NIN调控着ENOD11的空间表达, 并因此可能控制着结瘤数量; 而且, MtNIN对于自激活CCaMK诱导的自发结瘤过程是必需的。尽管hcl (hair curling)突变株与Mtnin有着相似的表型, 但HCL对于CCaMK诱导的根瘤形成却并非必不可少(Marsh et al., 2007)。研究表明, HCL编码的LYK3受体蛋白与日本百脉根中的NFR1同源, 也含有LysM结构域, LYK3控制着根毛卷曲, 并以依赖于结瘤因子结构的方式控制着侵入线的形成, 可能是结瘤因子的进入受体(Limpens et al., 2003; Smit et al., 2007)。
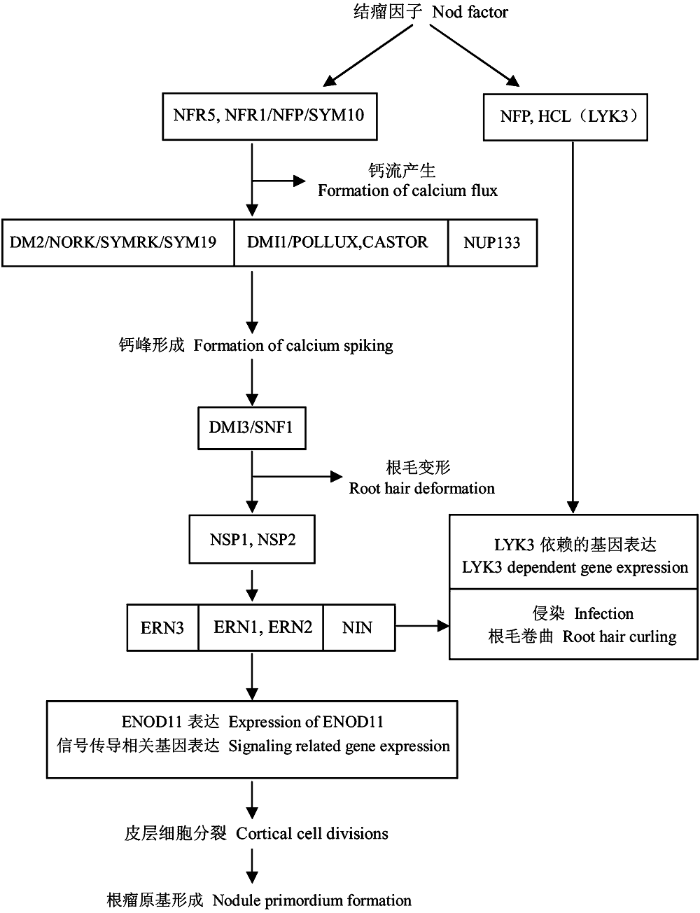
综上所述, 可以将目前已知的豆科植物早期共生结瘤过程中的信号流及生理反应表示为图2。
图2
图2
共生结瘤早期结瘤因子的信号传导途径。
Fig. 2
The Nod factor signaling pathways during early sym- biotic nodulation.
3 侵入线的形成及固氮的分子基础
这方面的研究主要通过Fix-突变株揭示, 该类突变株虽可结瘤或形成瘤状物, 但不能固氮。从蒺藜苜蓿中分离到了不少这种类型的突变株, 如形成小的、肿块状的无效根瘤的lin、nip、Mtsym1。lin (lumpy infections)只能形成根瘤原基, 侵染过程仅限于根表皮细胞, 皮层细胞中没有侵入线的形成, 接种后的根中检测不到乙炔还原活性(Kuppusamy et al., 2004)。nip (numerous infections and polyphenolics)虽然有侵入线的形成, 但发育很不正常, 且极少释放根瘤菌至宿主细胞或根瘤菌不能完成分化, 一些与植物防御有关的基因表达增强(Veere- shlingam et al., 2004)。在Mtsym1 (Bénaben et al., 1995)中, 晚期结瘤素基因如LB1、CAM1、N31不能表达, 也没有乙炔还原活性。最近, Teillet等(2008)从蒺藜苜蓿中分离到一个api (altered nodule primordium invasion)突变株, 表现为根瘤菌的侵染过程刚好在进入根瘤原基之前失败, 结果在与根瘤原基相邻的外皮层中形成大的侵入袋(侵入线变形增大后形成的袋状结构), 在根上形成一些小的瘤状物。研究显示, API与乙烯代谢及结瘤因子信号传递没有关系; 遗传图谱及上位分析显示, API应在ERN1和LIN的下游及NIP的上游行使功能。
从蒺藜苜蓿中, Starker等(2006)还筛选到了8个dnf (defective in nitrogen fixation)突变株, 它们均结白色小瘤。其中, 除了dnf3、dnf6可显示少量乙炔还原活性外, 其余突变株均检测不到固氮酶活性; 包括LB1、CAM1、N31在内的晚期结瘤素基因在大多数dnf突变株中都能正常表达, 只有dnf1、dnf5、dnf2例外: dnf1、dnf5不表达所有上述基因, dnf2则不表达N31。这些突变株中根瘤菌基因nodF (与侵染起始及根瘤发育有关)、exoY (与侵染有关)、bacA (为根瘤菌在宿主细胞中的存活所需)和nifH (固氮基因)的表达结果显示, dnf2中只有少量根瘤支持表达nodF和bacA; dnf1、dnf2和dnf5不能启动nifH的表达, 其余突变株表达nifH的根瘤数量只有野生型的26% (dnf4)、32% (dnf7)或一半(dnf3、dnf6); exoY在dnf3、dnf5、dnf6中的表达显著下降。可见, 一个有功能的根瘤不仅已经完成了组织分化, 还需要为根瘤菌提供合适的环境, 激活相关基因的表达, 才能发挥其固氮功能。
另外, 来自日本百脉根的crinkle (Ljsym79)突变导致侵入线不能穿过表皮细胞, 少数根瘤含有形状不规则的能固氮的类菌体, 能够表达豆血红蛋白基因; 植物本身也出现了异常发育(Tansengco et al., 2003)。在alb1 (Ljsym74)突变株形成的无效瘤中, 根瘤中的维管束发育不全, 根瘤菌被包裹在异常增大了的侵入线中, 不能释放进入植物细胞(Imaizumi- Anraku et al., 2000)。
4 豆科植物结瘤的自主调控分子机制
根瘤的形成虽然有利于植物在氮素营养不足的环境中生存, 但这毕竟是一个耗能的过程, 根瘤数目过多将对植物的生长造成不利的影响, 为了解决这一矛盾, 豆科植物在长期进化中已经发展出一套负反馈调节机制: 已经形成的根瘤能够抑制新的根瘤的形成。豆科植物的这一自主调控机制(autoregulation of nodulation, AON)主要通过根-茎-根之间的长距离信号交流实现: 受结瘤因子刺激后, 根部产生某种信号分子被运至茎部, 茎解读此信号后合成出新的信号分子再输送到根部, 从而对根瘤的数目进行控制。除此之外, 我们认为, AON调控体系还应该包括细胞-细胞间的短距离信号交流。另外, AON调控体系不仅控制着根瘤的数目, 同时也可能控制着根瘤的类型。
4.1 长距离交流体系中根产生的信号分子
日本百脉根超结瘤突变株har1-与野生型植株之间的嫁接试验表明, 超结瘤表型由茎的基因型决定, 从而引出茎产生的某种物质对根瘤数量进行控制的推测。序列分析显示, HAR1 (hypernodulation aberrant root formation 1)编码一个富含亮氨酸的类受体激酶, 和来自拟南芥(Arabidopsis thaliana)的CLAVATA1 (CLV1)有着很高的同源性(Krusell et al., 2002; Nishimura et al., 2002a)。CLV1是一个类受体激酶, 在拟南芥中, 通过与经由CLV3加工而来的小分子多肽相结合, 以细胞-细胞的短距离信号传递方式, 调控着茎和花顶端分生组织细胞的增殖(Ogawa et al., 2008)。CLV3属于CLE家族(CLAVATA3/ESR-related family)成员, 研究表明, CLE家族所编码的蛋白或加工后的小分子多肽常常作为胞外多肽激素发挥作用。此外, 大豆、豌豆和蒺藜苜蓿的同源基因产物NARK (nodule autoreg- ulation receptor kinase; Searle et al., 2003)、SYM29 (Krusell et al., 2002)、SUNN (super numeric nodules; Schnabel, et al., 2005)对根瘤数量的控制也都是通过茎-根之间的长距离信息交流实现的, 这些AON基因均与CLV1有着极高的序列相似性。因此, 有理由认为, AON信号调控系统也可能以CLE蛋白作为多肽激素, 激活了HAR1类受体激酶。
近年, 从日本百脉根中分离到2个CLE基因: LjCLE-RS1和LjCLE-RS2, 研究表明, 这两个基因都只在根中表达, 并在接种根瘤菌后的24 h内表达量迅速上升; 发根转化实验结果显示, 它们在野生型植株根中的过度表达强烈抑制了根瘤的形成, 而且, 这种抑制效果还能传递至未转化的根; 同时, HAR1这一类受体激酶对于LjCLE-RS1/RS2发挥抑制作用必不可少(Okamoto et al., 2009)。此外, 在蒺藜苜蓿中也发现了两个与根瘤数目控制有关的基因: RALFL1 (rapid alkalinization factor-like 1)和DVL1 (devil 1), 与LjCLE-RS1/RS2类似, 二者的超表达同样引起根瘤数目的急剧下降(Combier et al., 2008a)。但是, 这两个基因是否也作为AON基因行使功能, 尚待进一步研究。已经证实, 乙烯能够以局部调控的方式控制根瘤数目, 这种抑制作用不同于AON的系统控制。蒺藜苜蓿的乙烯不敏感突变体sickle, 也表现过度结瘤现象, 且具备完整的AON调控系统; 其中的相关基因MtSkl1与拟南芥中的乙烯信号传导基因Ein2同源(Penmetsa et al., 2008)。因此, 也可能与MtSkl1类似, MtRALFL1和MtDVL1介导了结瘤因子与乙烯调控系统之间的某种分子 对话。
4.2 长距离交流体系中茎对来自根的信号分子的响应
已有研究表明, LjHAR1的启动子主要在叶、茎, 以及根瘤组织中的韧皮部表达(Combier et al., 2008a)。而韧皮部被普遍认为是长距离信号传递通道(Lough & Lucas, 2006)。鉴于此, Magori和Kawaguch (2009)认为, 来自根部的小分子多肽如CLE-RS1/2被输送到茎部, 直接或间接地激活了CLV1类受体激酶, 而CLV1的主要作用可能是介导茎中合成相应的信号分子并与之结合, 随后通过韧皮部运至根部, 并与相应的受体结合。然而, 茎中合成的信号分子究竟是什么, 目前尚无确切的报道, 尽管有一些研究表明, 茉莉酸、生长素等植物激素可能在AON系统中发挥作用, 但实验结果往往相互矛盾。此外, 类黄酮也可能通过介导生长素的极性运输而影响根瘤的形成(Hirsch, 1992)。在蒺藜苜蓿和大豆中, 类黄酮合成途径中相关基因的表达沉默导致结瘤减少, 同时增强了生长素由茎到根的运输水平(Subramanian et al., 2006; Wasson et al., 2006)。但是, 在大豆中, 类黄酮合成酶基因的沉默导致的结瘤下降可以通过加入结瘤因子等方法解除(Subramanian et al., 2006), 说明至少在大豆中, 类黄酮对结瘤过程的影响也许并不是通过对生长素运输的调控实现的。
虽然来自茎的因子尚未证实, 但其在根中的可能受体或信号中间载体已经被报道: 嫁接试验表明, 与har1不同, 日本百脉根突变株tml (too much love)超结瘤表型是由根的基因型控制的, 而且har1突变不能使tml的根瘤数目明显增加; 此外, TML对结瘤的抑制作用不能在根与根之间传递。因此, TML可能是茎信号分子的根部受体, 在har1下游起作用(Magori et al., 2009)。考虑到AON的系统调控方式, 显然, 在TML下游应该有可移动信号分子的产生, 以使茎对根瘤形成的抑制效应能在根与根之间传递。
除了CLV1类受体激酶, KLAVIER(KLV)是另一个自日本百脉根的茎中获得的AON信号体系成员。但是, klavier突变株除了根瘤数目增多外, 还出现叶脉凸起、开花延迟、植株矮化等现象(Oka-Kira et al., 2005)。日本百脉根的另一突变株astray, 能形成二倍于野生型的根瘤, 且提早结瘤, 在非共生条件下, 此突变株对重力和光的反应存在缺陷(Nishimura et al., 2002b)。ASTRAY能够在植物的叶片、子叶、茎、胚轴、根及根瘤等组织中表达, 根部是否接种根瘤菌对其表达没有明显影响。ASTRAY编码蛋白质LjBzf, 其C端具有亮氨酸锌指结构域, 与拟南芥HY5蛋白高度同源, HY5是一个与光形态发生有关的调控因子; LjBzf的N端含有一个RING- finger和一个酸性区域。Nishimura等(2002b)认为, LjBzf中包含的结构域组合为豆科植物所特有的, 负控制着根瘤的形成。这些现象说明, KLV和ASTRAY可能是植物生长发育过程中一个多功能的普遍性调控因子, 不仅控制着根瘤的形成, 同时还调控着植物的其他生理过程。
4.3 细胞与细胞之间短距离交流对根瘤形成过程的控制
实际上, 除了茎与根之间长距离信号传递体系外, 根瘤起始和根瘤数目的调控应该还包括细胞与细胞之间的短距离分子对话, 后者可能主要以胞间连丝作为交流通道(plasmodesmata, PD)。通过荧光跟踪、绿色荧光蛋白(GFP)报告系统等方法, 研究人员分析了蒺藜苜蓿中PD介导的韧皮部与根瘤原基之间的信息交流情况(Imlau et al., 1999; Complainville et al., 2003)。研究发现, 接种根瘤菌后, 在刚刚开始分裂的皮层细胞中, 即可检测到GFP信号, 而根瘤原基正是由这些皮层细胞分裂形成的。在这些根瘤起始细胞中, GFP在细胞分裂完成前即已表达, 这与PD网络重新排布相一致, 说明PD介导了NF激活后的植物细胞与其他部分之间的分子交流, 这对根瘤的形成可能非常重要(Complainville et al., 2003)。AON体系中最终能在根与根之间进行某种抑制因子的传递, 可能正是经由PD的短距离交流实现的。
4.4 茎对根瘤类型的控制
茎不但控制着结瘤数量, 而且在决定根瘤的类型上也起着重要作用。通常情况下, 蒺藜苜蓿与Sinorhizobium meliloti共生形成不定型根瘤, 而日本百脉根与Mesorhizobium loti共生形成定型根瘤。两者之间的嫁接试验表明, 嫁接了蒺藜苜蓿茎的日本百脉根的根能与M. loti共生形成定型根瘤, 它的超结瘤突变株har1-1的根嫁接到蒺藜苜蓿茎上后可恢复野生型结瘤现象; 然而, 当蒺藜苜蓿的根与日本百脉根的茎嫁接后, 无论接种S. meliloti还是M. loti, 都不能结瘤, 说明日本百脉根的茎不能支持蒺藜苜蓿的根与相应根瘤菌之间的结瘤, 不定型根瘤的形成也许还需要某种来自茎的因子的参与(Lohar & VandenBosch, 2005)。茎对根瘤类型的控制可能也是AON体系的一部分。
5 环境中的氮素营养对根瘤形成的影响
豆科植物根瘤的形成不仅涉及共生双方的分子对话与调控, 同时也与环境因素有关, 如水分、矿质营养元素、温度、重金属、钠盐、CO2、土壤类型以及pH等, 其中, 研究较为深入的是结合态氮素的影响。土壤中氮素养料的匮乏是根瘤形成、发育乃至发挥固氮功能必不可少的先决条件, 高浓度的硝酸盐或铵盐的存在能够完全抑制根瘤的形成(Oka-kira et al., 2005; Barbulova et al., 2007), 而低浓度的氮能促进植物生长并促进结瘤(Fei & Vessey, 2009)。实际上, 来自不同豆科植物的超结瘤突变株都表现出了对硝酸盐的耐受性(Krusell et al., 2002; Nishimura et al., 2002a; Searle et al., 2003; Schnabel et al., 2005)。在日本百脉根中, 结合态氮的存在抑制了NF诱导的皮层细胞的分裂; 相对于生长在无氮条件下的植株, 在高浓度硝酸盐或铵盐环境中生长的野生型日本百脉根, 在受NF诱导或接种M. loti 24 h后仍无NIN基因的表达; 对于har1-3超结瘤突变株, 即使在培养基中添加10或20 mmol∙L-1 KNO3的情况下, NIN仍能被高水平诱导, 但是, 当加入10 mmol∙L-1 NH4NO3或琥珀酸铵时, NIN的表达与野生型一样被抑制。说明硝态氮和铵态氮可能在根瘤原基形成的不同时期发挥抑制作用, 而NIN则在硝态氮的抑制信号途径中起关键作用(Barbulova et al., 2007)。此外, 硝酸盐增强了LjCLE-RS2在植物根中的表达, 因此, har1超结瘤突变株之所以在有硝酸盐的环境中仍然结瘤, 可能正是因为与硝酸盐信号途径相关的LjCLE-RS2失去了识别受体(Okamoto et al., 2009)。
6 展望
6.1 信号传递相关基因的分离与根瘤形成的基因调控网络的完善
共生固氮分子机制涉及到宿主植物对细菌结瘤因子的特异性识别及宿主结瘤素基因的表达激活, 其中包括与信号传导有关的基因, 例如相关激酶和转录因子的表达。目前, 对宿主植物对NF早期反应的分析、NF受体的克隆及部分信号传导基因的分离, 已经在共生互作与植物信号传递之间打开了一道门。尽管已从不同的豆科植物中分离到不少结瘤素基因, 但是, 大部分基因在结瘤过程中的具体作用并不完全清楚。例如, 大量的编码根瘤特异性的富含半胱氨酸簇的蛋白质(cysteine cluster protein, CCP)基因在不定型根瘤中被分离(Mergaert et al., 2003; Chou et al., 2006), 这些CCP具有抗微生物的保护蛋白的结构域, 它们在根瘤中的转录明显增加, 其生物学功能尚待研究。从上面的介绍中也可以看到, 迄今分离到的与根瘤形成有关的转录因子, 如NSP、ERN、NIN等, 在根瘤形成中的具体功能需要进一步证实。而且, 目前对类似的调控根瘤形成的结瘤素基因仍然知之甚少。无疑, 包括植物激素在内的共生早期信号分子的分离及其功能研究, 仍将继续是共生领域的研究热点; 而随后的共生过程, 如侵入线形成、根瘤菌的进入与释放等现象的探明也非常令人期待, 这将为最终建立一个完整明确的根瘤形成的基因调控网络提供依据。
6.2 转录因子与小分子RNAs的作用
基因的转录动力学不仅与转录过程本身有关, 而且与mRNA的稳定性相关, 特别是在真核细胞中, mRNA的稳定性与细胞分化有着重要关系, 并受到不编码的小分子RNAs (microRNAs, miRNAs)的调控。许多转录因子组成复杂的调控网络, 负责器官发生, 根瘤的形成也不例外。在植物细胞中, miRNAs调节着转录因子的活性, 编码转录因子的mRNAs正是miRNAs的作用目标(Brodersen et al., 2008)。根瘤的形成中, 也发现了类似的调控方式。来自蒺藜苜蓿的MtHAP2.1是目前分离到的少数在根瘤分生组织细胞中表达的基因之一, 编码CCAAT结合蛋白家族的转录因子, 其表达在根瘤的顶端分生组织中最为丰富, 在与之毗邻的侵染带的细胞中表达降低, 而在根瘤的其他组织中均检测不到。研究显示, MtHAP2.1的表达受到microRNA- 169 (MIR169)的调控, MIR169的过度表达与利用MtHAP2.1 RNAi的实验结果相似, 都表现为根瘤的发育受阻; 而MIR169介导的转录后调控功能的丧失则导致根瘤发育迟缓(Combier et al., 2006, 2008b)。因此, MIR169可能通过限定MtHAP2.1的空间表达以及快速的转录后降解来促使根瘤细胞分化。在蒺藜苜蓿中, 另一个小分子RNA――MIR166能够对HD-ZipIII (Class-III homeodomain-leucine zipper)家族的转录因子进行转录后调控, 后者在根的维管组织中表达并与根瘤形成有关。MIR166的过度表达至少降低了3个HD-Zip III家族成员的转录, 并因此影响了根维管组织的发育, 阻碍了根瘤和侧根的形成起始(Boualem et al., 2008)。可见, 在根瘤器官发生的起始分化和根瘤细胞形成过程中, miRNAs调控着其间涉及的转录因子的时空表达方式。在不久的将来, 可能会有更多的与根瘤形成有关的小分子RNAs被鉴定。因此, 更多转录因子及其靶位启动子的证实和生物学活性的探明, miRNAs的分离及其功能研究, 特别是豆科植物特异性的miRNAs的研究, 应是未来豆科植物-根瘤菌共生互作领域的一个研究热点。
6.3 根瘤形成的进化研究
前文述及的参与根瘤形成的激酶、转录因子等调控基因同时也参与了侧根的形成, 或者与植物和菌根真菌的共生互作有关, 后者是一个植物王国中更为广泛、更为古老的共生体系; 并且, 已经分离到的大多数结瘤素基因, 包括调控基因在内, 都能在非豆科植物中找到同源序列, 其中包括激酶、转录因子等参与调控的基因。Szczyglowski和Amyot (2003)认为, 在进化过程中, 豆科植物可能是通过利用植物业已存在的发育调控机制或其中的调控因子建立起与根瘤菌的共生关系的。因此, 不难想象, 在豆科植物与根瘤菌的共生互作中, 植物可能通过对根形态发生的调控方式的重新调整, 而不是或不完全是通过产生新的蛋白质来促使根瘤形成。豆科植物之间、豆科植物与非豆科植物之间比较基因组学的研究将有助于解答这个问题。
6.4 AON信号途径的明确及其对环境因子的响应
根瘤的形成是共生体系内部的信息交流及其与环境因子相互作用的结果, 这就需要一个能将内、外信号链接起来的系统调控方式。与NF信号体系相比, AON信号途径正是这样一个不仅能对共生体系内部信号(如NF)作出反应, 同时还能对外界环境因子(如氮素浓度)作出回应的系统调控体系。某些环境因子(如氮素营养)对共生结瘤的影响也许就是通过AON实现的。然而, 目前关于AON的研究才刚刚起步, 其信号传递的具体途径仍不明确, 这需要进一步分离AON相关基因并对其功能进行研究。同时, 环境因子影响根瘤发育过程的分子机制问题, 也是目前共生固氮研究领域里的一个薄弱环节。应用反向遗传学手段, 将AON信号传递体系与环境因子, 特别是不同结合态氮素对结瘤的影响相结合, 将更有利于揭示这一豆科植物特有的自主调控方式。
致谢
陕西省自然科学基金(2009JM3011)和西北农林科技大学基本科研业务费专项资金(QN2009068)资助项目。华中农业大学农业微生物学国家重点实验室生物固氮研究室给予支持, 在此表示感谢。
参考文献
The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation
AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif
Rhizobium Nod factors (NFs) are specific lipochitooligosaccharides that activate host legume signaling pathways essential for initiating the nitrogen-fixing symbiotic association. This study describes the characterization of cis-regulatory elements and trans-interacting factors that regulate NF-dependent and epidermis-specific gene transcription in Medicago truncatula. Detailed analysis of the Mt ENOD11 promoter using deletion, mutation, and gain-of-function constructs has led to the identification of an NF-responsive regulatory unit (the NF box) sufficient to direct NF-elicited expression in root hairs. NF box-mediated expression requires a major GCC-like motif, which is also essential for the binding of root hair-specific nuclear factors. Yeast one-hybrid screening has identified three closely related AP2/ERF transcription factors (ERN1 to ERN3) that are able to bind specifically to the NF box. ERN1 is identical to an ERF-like factor identified recently. Expression analysis has revealed that ERN1 and ERN2 genes are upregulated in root hairs following NF treatment and that this activation requires a functional NFP gene. Transient expression assays in Nicotiana benthamiana have further shown that nucleus-targeted ERN1 and ERN2 factors activate NF box-containing reporters, whereas ERN3 represses ERN1/ERN2-dependent transcription activation. A model is proposed for the fine-tuning of NF-elicited gene transcription in root hairs involving the interplay between repressor and activator ERN factors.
Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes
Legumes form symbiotic associations with both mycorrhizal fungi and nitrogen-fixing soil bacteria called rhizobia. Several of the plant genes required for transduction of rhizobial signals, the Nod factors, are also necessary for mycorrhizal symbiosis. Here, we describe the cloning and characterization of one such gene from the legume Medicago truncatula. The DMI1 (does not make infections) gene encodes a novel protein with low global similarity to a ligand-gated cation channel domain of archaea. The protein is highly conserved in angiosperms and ancestral to land plants. We suggest that DMI1 represents an ancient plant-specific innovation, potentially enabling mycorrhizal associations.
Rhizobium meliloti lipooligosaccharide nodulation factors, different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses
Rhizobium meliloti produces lipochitooligosaccharide nodulation NodRm factors that are required for nodulation of legume hosts. NodRm factors are O-acetylated and N-acylated by specific C16-unsaturated fatty acids. nodL mutants produce non-O-acetylated factors, and nodFE mutants produce factors with modified acyl substituents. Both mutants exhibited a significantly reduced capacity to elicit infection thread (IT) formation in alfalfa. However, once initiated, ITs developed and allowed the formation of nitrogen-fixing nodules. In contrast, double nodF/nodL mutants were unable to penetrate into legume hosts and to form ITs. Nevertheless, these mutants induced widespread cell wall tip growth in trichoblasts and other epidermal cells and were also able to elicit cortical cell activation at a distance. NodRm factor structural requirements are thus clearly more stringent for bacterial entry than for the elicitation of developmental plant responses.
A positive regulatory role for LjERF1 in the nodulation process is revealed by systematic analysis of nodule-associated transcription factors of Lotus japonicus
We have used reverse genetics to identify genes involved in legume-rhizobium symbiosis in Lotus japonicus. We obtained the sequences of 20 putative transcription factors from previously reported large-scale transcriptome data. The transcription factors were classified according to their DNA binding domains and patterns of expression during the nodulation process. We identified two homologues of Medicago truncatula MtHAP2-1, which encodes a CCAAT-binding protein and has been shown to play a role in nodulation. The functions of the remaining genes in the nodulation process have not been reported. Seven genes were found to encode proteins with AP2-EREBP domains, six of which were similar to proteins that have been implicated in ethylene and/or jasmonic acid signal transduction and defense gene regulation in Arabidopsis (Arabidopsis thaliana). We identified a gene, LjERF1, that is most similar to Arabidopsis ERF1, which is up-regulated by ethylene and jasmonic acid and activates downstream defense genes. LjERF1 showed the same pattern of up-regulation in roots as Arabidopsis ERF1. The nodulation phenotype of roots that overexpressed LjERF1 or inhibited LjERF1 expression using an RNA interference construct indicated that this gene functions as a positive regulator of nodulation. We propose that LjERF1 functions as a key regulator of successful infection of L. japonicus by Mesorhizobium loti.
Divergence of evolutionary ways among common sym genes, CASTOR and CCaMK show functional conservation between two symbiosis systems and constitute the root of a common signaling pathway
In recent years a number of legume genes involved in root nodule (RN) symbiosis have been identified in the model legumes, Lotus japonicus (Lotus) and Medicago truncatula. Among them, a distinct set of genes has been categorized as a common symbiosis pathway (CSP), because they are also essential for another mutual interaction, the arbuscular mycorrhiza (AM) symbiosis, which is evolutionarily older than the RN symbiosis and is widely distributed in the plant kingdom. Based on the concept that the legume RN symbiosis has evolved from the ancient AM symbiosis, one issue is whether the CSP is functionally conserved between non-nodulating plants, such as rice, and nodulating legumes. We identified three rice CSP gene orthologs, OsCASTOR, OsPOLLUX and OsCCaMK, and demonstrated the indispensable roles of OsPOLLUX and OsCCaMK in rice AM symbiosis. Interestingly, molecular transfection of either OsCASTOR or OsCCaMK could fully complement symbiosis defects in the corresponding Lotus mutant lines for both the AM and RN symbioses. Our results not only provide a conserved genetic basis for the AM symbiosis between rice and Lotus, but also indicate that the core of the CSP has been well conserved during the evolution of RN symbiosis. Through evolution, CASTOR and CCaMK have remained as the molecular basis for the maintenance of CSP functions in the two symbiosis systems.
Differential effects of combined N sources on early steps of the Nod factor-dependent transduction pathway in Lotus japonicus
TE7, an inefficient symbiotic mutant of Medicago truncatula
A mutagenesis program using ethylmethane sulfonate on Medicago truncatula Gaertn cv Jemalong, an annual, autogamous and diploid lucerne, permitted the isolation of a mutant (TE7) unable to establish an effective nitrogen-fixing symbiosis, [Nod+Fix-], with Rhizobium meliloti wild-type strains. The mutant phenotype is characterized by an altered infection process that leads to the formation of two kinds of inefficient nodules on the same root system. A certain proportion of the nodules are small, round, and uninfected, with infection threads limited to the outer root cortical cells. Others develop to a normal elongated shape and are infected; bacterial release occurs but the bacteria do not differentiate into bacteroids. The ratio of invaded to uninvaded nodules depends on the bacterial strain used. Throughout the infection process, certain events correlated with the plant defense response against pathogens can be observed: (a) the presence of polyphenolic compounds associated with the walls of infected cells and also with some parts of infection threads in the root cortex; (b) appositions on infection thread walls during the early stage of infection and also within the central tissue of infected nodules; and (c) autophagy of the plant cells that contain released bacteria. Genetic data suggest that the phenotype of TE7 is under monogenic and recessive control; this gene has been designated Mtsym1.
The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus
MicroRNA166 controls root and nodule development in Medicago truncatula
Development of the legume root nodule
Tissue and cell invasion by Rhizobium, the structure and development of infection threads and symbiosomes
Widespread translational inhibition by plant miRNAs and siRNAs
High complementarity between plant microRNAs (miRNAs) and their messenger RNA targets is thought to cause silencing, prevalently by endonucleolytic cleavage. We have isolated Arabidopsis mutants defective in miRNA action. Their analysis provides evidence that plant miRNA-guided silencing has a widespread translational inhibitory component that is genetically separable from endonucleolytic cleavage. We further show that the same is true of silencing mediated by small interfering RNA (siRNA) populations. Translational repression is effected in part by the ARGONAUTE proteins AGO1 and AGO10. It also requires the activity of the microtubule-severing enzyme katanin, implicating cytoskeleton dynamics in miRNA action, as recently suggested from animal studies. Also as in animals, the decapping component VARICOSE (VCS)/Ge-1 is required for translational repression by miRNAs, which suggests that the underlying mechanisms in the two kingdoms are related.
SrSymRK, a plant receptor essential for symbiosome formation
The symbiosis between legumes and rhizobia is essential for the nitrogen input into the life cycle on our planet. New root organs, the nodules, are established, which house N2-fixing bacteria internalized into the host cell cytoplasm as horizontally acquired organelles, the symbiosomes. The interaction is initiated by bacterial invasion via epidermal root hair curling and cell division in the cortex, both triggered by bacterial nodulation factors. Of the several genes involved in nodule initiation that have been identified, one encodes the leucine-rich repeat-type receptor kinase SymRK. In SymRK mutants of Lotus japonicus or its orthologs in Medicago sp. and Pisum sativum, nodule initiation is arrested at the level of the root hair interaction. Because of the epidermal block, the role of SymRK at later stages of nodule development remained enigmatic. To analyze the role of SymRK downstream of the epidermis, the water-tolerant legume Sesbania rostrata was used that has developed a nodulation strategy to circumvent root hair responses for bacterial invasion. Evidence is provided that SymRK plays an essential role during endosymbiotic uptake in plant cells.
Ion changes in legume root hairs responding to Nod factors
Antiquity and function of CASTOR and POLLUX, the twin ion channel-encoding genes key to the evolution of root symbioses in plants
Thirteen nodule-specific or nodule-enhanced genes encoding products homologous to cysteine cluster proteins or plant lipid transfer proteins are identified in Astragalus sinicus L. by suppressive subtractive hybridization
Thirteen nodule-specific or nodule-enhanced genes have been revealed by suppressive subtractive hybridization (SSH) with two mRNA populations of infected and uninfected control roots of Astragalus sinicus. Eleven of them encode small polypeptides showing homology to cysteine cluster proteins (CCPs) that contain a putative signal peptide and conserved cysteine residues. Among these CCP-like genes, AsG257 codes for a homologue of the defensin 2 family and AsD255 contains a scorpion toxin-like domain at the C-terminus. Sequence analysis of a genomic AsD255 fragment which was isolated revealed that one intron separates the first exon encoding the signal peptide from the second exon encoding the cysteine cluster domain of this nodulin. Another two genes, AsE246 and AsIB259, encode two different products similar to lipid transfer proteins (LTPs). Virtual northern blot and reverse transcription-polymerase chain reaction (RT-PCR) analysis indicated that the other genes except AsIB259 and AsC2411 were expressed exclusively in inoculated roots and that their expression was 2-4 d later than that of the leghaemoglobin (Lb) gene during nodule development. Transcription of AsIB259 was also detected in uninfected control roots but with a significant decline in expression and a temporal expression similar to Lb. AsC2411 had a basal expression in control roots identified by RT-PCR. Sequence alignment showed that the putative proteins AsE246 and AsIB259 show lower homology with LTPs from legumes than with those from other plants.
Evidence for the involvement in nodulation of the two small putative regulatory peptide-encoding genes MtRALFL1 and MtDVL1
Nod factors are key bacterial signaling molecules regulating the symbiotic interaction between bacteria known as rhizobia and leguminous plants. Studying plant host genes whose expression is affected by Nod factors has given insights into early symbiotic signaling and development. Here, we used a double supernodulating mutant line that shows increased sensitivity to Nod factors to study the Nod factor-regulated transcriptome. Using microarrays containing more than 16,000 70-mer oligonucleotide probes, we identified 643 Nod-factor-regulated genes, including 225 new Nod-factor-upregulated genes encoding many potential regulators. Among the genes found to be Nod factor upregulated, we identified and characterized MtRALFL1 and MtDVL1, which code for two small putative peptide regulators of 135 and 53 amino acids, respectively. Expression analysis confirmed that these genes are upregulated during initial phases of nodulation. Overexpression of MtRALFL1 and MtDVL1 in Medicago truncatula roots resulted in a marked reduction in the number of nodules formed and in a strong increase in the number of aborted infection threads. In addition, abnormal nodule development was observed when MtRALFL1 was overexpressed. This work provides evidence for the involvement of new putative small-peptide regulators during nodulation.
Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development
MtHAP2-1 is a CCAAT-binding transcription factor from the model legume Medicago truncatula. We previously showed that MtHAP2-1 expression is regulated both spatially and temporally by microRNA169. Here we present a novel regulatory mechanism controlling MtHAP2-1 expression. Alternative splicing of an intron in the MtHAP2-1 5'leader sequence (LS) becomes predominant during the development of root nodules, leading to the production of a small peptide, uORF1p. Our results indicate that binding of uORF1p to MtHAP2-1 5'LS mRNA leads to reduced accumulation of the MtHAP2-1 transcript and may contribute to spatial restriction of MtHAP2-1 expression within the nodule. We propose that miR169 and uORF1p play essential, sequential, and nonredundant roles in regulating MtHAP2-1 expression. Importantly, in contrast to previously described cis-acting uORFs, uORF1p is able to act in trans to down-regulate gene expression. Our work thus contributes to a better understanding of the action of upstream ORFs (uORFs) in the regulation of gene expression.
Nodule initiation involves the creation of a new symplasmic field in specific root cells of Medicago species
The organogenesis of nitrogen-fixing nodules in legume plants is initiated in specific root cortical cells and regulated by long-distance signaling and carbon allocation. Here, we explore cell-to-cell communication processes that occur during nodule initiation in Medicago species and their functional relevance using a combination of fluorescent tracers, electron microscopy, and transgenic plants. Nodule initiation induced symplasmic continuity between the phloem and nodule initials. Macromolecules such as green fluorescent protein could traffic across short or long distances from the phloem into these primordial cells. The created symplasmic field was regulated throughout nodule development. Furthermore, Medicago truncatula transgenic plants expressing a viral movement protein showed increased nodulation. Hence, the establishment of this symplasmic field may be a critical element for the control of nodule organogenesis.
Rhizobium lipo- chitooligosaccharide nodulation factors, signaling molecules mediating recognition and morphogenesis
Rhizobium nodulation signals
A receptor kinase gene regulating symbiotic nodule development
Stimulation of nodulation in Medicago truncatula by low concentrations of ammonium: quantitative reverse transcription PCR analysis of selected genes
Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition
Legumes, such as Medicago truncatula, form mutualistic symbiotic relationships with nitrogen-fixing rhizobial bacteria. This occurs within specialized root organs--nodules--that provide the conditions required for nitrogen fixation. A rhizobium-derived signalling molecule, Nod factor, is required to establish the symbiosis. Perception of Nod factor in the plant leads to the induction of Ca2+ oscillations, and the transduction of this Ca2+ signal requires DMI3 (refs 2, 3), which encodes the protein kinase Ca2+/calmodulin-dependent protein kinase (CCaMK). Central to the regulation of CCaMK is an autoinhibitory domain that negatively regulates kinase activity. Here we show that the specific removal of the autoinhibition domain leads to the autoactivation of the nodulation signalling pathway in the plant, with the resultant induction of nodules and nodulation gene expression in the absence of bacterial elicitation. This autoactivation requires nodulation-specific transcriptional regulators in the GRAS family. This work demonstrates that the release of autoinhibition from CCaMK after calmodulin binding is a central switch that is sufficient to activate nodule morphogenesis. The fact that a single regulation event is sufficient to induce nodulation highlights the possibility of transferring this process to non-legumes.
Legumes, importance and constraints to greater use
Developmental biology of legume nodulation
GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula
The symbiotic association of legumes with rhizobia involves bacterially derived Nod factor, which is sufficient to activate the formation of nodules on the roots of the host plant. Perception of Nod factor by root hair cells induces calcium oscillations that are a component of the Nod factor signal transduction pathway. Perception of the calcium oscillations is a function of a calcium- and calmodulin-dependent protein kinase, and this activates nodulation gene expression via two GRAS domain transcriptional regulators, Nodulation Signaling Pathway1 (NSP1) and NSP2, and an ERF transcription factor required for nodulation. Here, we show that NSP1 and NSP2 form a complex that is associated with the promoters of early nodulin genes. We show that NSP1 binds directly to ENOD promoters through the novel cis-element AATTT. While NSP1 shows direct binding to the ENOD11 promoter in vitro, this association in vivo requires NSP2. The NSP1-NSP2 association with the ENOD11 promoter is enhanced following Nod factor elicitation. Mutations in the domain of NSP2 responsible for its interaction with NSP1 highlight the significance of the NSP1-NSP2 heteropolymer for nodulation signaling. Our work reveals direct binding of a GRAS protein complex to DNA and highlights the importance of the NSP1-NSP2 complex for efficient nodulation in the model legume Medicago truncatula.
Analysis of ENOD40 expression in alb1, a symbiotic mutant of Lotus japonicus that forms empty nodules with incompletely developed nodule vascular bundles
The alb1 mutant of Lotus japonicus (Ljsym74) forms empty nodules in which most of the bacteria remain in abnormally enlarged infection threads and fail to enter the host plant cells. The alb1 mutant was also found to be defective in differentiation of ramified nodule vascular bundles; only a single vascular bundle differentiates at the proximal end of the alb1 nodules and it fails to differentiate further. Histochemical analysis using fluorescein-conjugated wheat-germ agglutinin (F-WGA) indicated that the mutation in the ALB1 gene specifically affects the differentiation of vascular bundles in nodules. Analysis of nodulin gene expression revealed that the expression of an early nodulin gene, ENOD40, was very low in alb1 nodules. At early developmental stages of alb1 nodules, the pattern of ENOD40 transcription was essentially the same as that in wild-type nodules; transcripts were localized in dividing cortical cells and in the pericycle of the root stele opposite nodule primordia, as in wild-type nodules. However, mature alb1 nodules exhibited very weak or no expression of ENOD40 in the peripheral cells of the undeveloped nodule vascular bundle. The ENOD40 expression pattern in alb1 nodules is distinct from that in another ineffective mutant, fen1 (Ljsym76), in which ENOD40 expression persists prior to premature senescence. These findings lead us to speculate that ENOD40 may play a role in the differentiation of nodule vascular bundles.
Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots
The roots of most higher plants form arbuscular mycorrhiza, an ancient, phosphate-acquiring symbiosis with fungi, whereas only four related plant orders are able to engage in the evolutionary younger nitrogen-fixing root-nodule symbiosis with bacteria. Plant symbioses with bacteria and fungi require a set of common signal transduction components that redirect root cell development. Here we present two highly homologous genes from Lotus japonicus, CASTOR and POLLUX, that are indispensable for microbial admission into plant cells and act upstream of intracellular calcium spiking, one of the earliest plant responses to symbiotic stimulation. Surprisingly, both twin proteins are localized in the plastids of root cells, indicating a previously unrecognized role of this ancient endosymbiont in controlling intracellular symbioses that evolved more recently.
Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues
Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators
Rhizobial bacteria enter a symbiotic interaction with legumes, activating diverse responses in roots through the lipochito oligosaccharide signaling molecule Nod factor. Here, we show that NSP2 from Medicago truncatula encodes a GRAS protein essential for Nod-factor signaling. NSP2 functions downstream of Nod-factor-induced calcium spiking and a calcium/calmodulin-dependent protein kinase. We show that NSP2-GFP expressed from a constitutive promoter is localized to the endoplasmic reticulum/nuclear envelope and relocalizes to the nucleus after Nod-factor elicitation. This work provides evidence that a GRAS protein transduces calcium signals in plants and provides a possible regulator of Nod-factor-inducible gene expression.
A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis
Shoot control of root development and nodulation is mediated by a receptor like kinase
In legumes, root nodule organogenesis is activated in response to morphogenic lipochitin oligosaccharides that are synthesized by bacteria, commonly known as rhizobia. Successful symbiotic interaction results in the formation of highly specialized organs called root nodules, which provide a unique environment for symbiotic nitrogen fixation. In wild-type plants the number of nodules is regulated by a signalling mechanism integrating environmental and developmental cues to arrest most rhizobial infections within the susceptible zone of the root. Furthermore, a feedback mechanism controls the temporal and spatial susceptibility to infection of the root system. This mechanism is referred to as autoregulation of nodulation, as earlier nodulation events inhibit nodulation of younger root tissues. Lotus japonicus plants homozygous for a mutation in the hypernodulation aberrant root (har1) locus escape this regulation and form an excessive number of nodules. Here we report the molecular cloning and expression analysis of the HAR1 gene and the pea orthologue, Pisum sativum, SYM29. HAR1 encodes a putative serine/threonine receptor kinase, which is required for shoot-controlled regulation of root growth, nodule number, and for nitrate sensitivity of symbiotic development.
LIN, a Medicago truncatula gene required for nodule differentiation and persistence of rhizobial infections
A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses
Legumes can enter into symbiotic relationships with both nitrogen-fixing bacteria (rhizobia) and mycorrhizal fungi. Nodulation by rhizobia results from a signal transduction pathway induced in legume roots by rhizobial Nod factors. DMI3, a Medicago truncatula gene that acts immediately downstream of calcium spiking in this signaling pathway and is required for both nodulation and mycorrhizal infection, has high sequence similarity to genes encoding calcium and calmodulin-dependent protein kinases (CCaMKs). This indicates that calcium spiking is likely an essential component of the signaling cascade leading to nodule development and mycorrhizal infection, and sheds light on the biological role of plant CCaMKs.
LysM domain receptor kinases regulating rhizobial Nod factor-induced infection
The rhizobial infection of legumes has the most stringent demand toward Nod factor structure of all host responses, and therefore a specific Nod factor entry receptor has been proposed. The SYM2 gene identified in certain ecotypes of pea (Pisum sativum) is a good candidate for such an entry receptor. We exploited the close phylogenetic relationship of pea and the model legume Medicago truncatula to identify genes specifically involved in rhizobial infection. The SYM2 orthologous region of M. truncatula contains 15 putative receptor-like genes, of which 7 are LysM domain-containing receptor-like kinases (LYKs). Using reverse genetics in M. truncatula, we show that two LYK genes are specifically involved in infection thread formation. This, as well as the properties of the LysM domains, strongly suggests that they are Nod factor entry receptors.
Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2
In most legume nodules, the N2-fixing rhizobia are present as organelle-like structures inside their host cells. These structures, named symbiosomes, contain one or a few rhizobia surrounded by a plant membrane. Symbiosome formation requires the release of bacteria from cell-wall-bound infection threads. In primitive legumes, rhizobia are hosted in intracellular infection threads that, in contrast to symbiosomes, are bound by a cell wall. The formation of symbiosomes is presumed to represent a major step in the evolution of legume-nodule symbiosis, because symbiosomes facilitate the exchange of metabolites between the two symbionts. Here, we show that the genes, which are essential for initiating nodule formation, are also actively transcribed in mature Medicago truncatula nodules in the region where symbiosome formation occurs. At least one of these genes, encoding the receptor kinase DOES NOT MAKE INFECTIONS 2 (DMI2) is essential for symbiosome formation. The protein locates to the host cell plasma membrane and to the membrane surrounding the infection threads. A partial reduction of DMI2 expression causes a phenotype that resembles the infection structures found in primitive legume nodules, because infected cells are occupied by large intracellular infection threads instead of by organelle-like symbiosomes.
Metabolism of Rhizobium bacteroids
Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis
The biological reduction of atmospheric N2 to ammonium (nitrogen fixation) provides about 65% of the biosphere's available nitrogen. Most of this ammonium is contributed by legume-rhizobia symbioses, which are initiated by the infection of legume hosts by bacteria (rhizobia), resulting in formation of root nodules. Within the nodules, rhizobia are found as bacteroids, which perform the nitrogen fixation: to do this, they obtain sources of carbon and energy from the plant, in the form of dicarboxylic acids. It has been thought that, in return, bacteroids simply provide the plant with ammonium. But here we show that a more complex amino-acid cycle is essential for symbiotic nitrogen fixation by Rhizobium in pea nodules. The plant provides amino acids to the bacteroids, enabling them to shut down their ammonium assimilation. In return, bacteroids act like plant organelles to cycle amino acids back to the plant for asparagine synthesis. The mutual dependence of this exchange prevents the symbiosis being dominated by the plant, and provides a selective pressure for the evolution of mutualism.
Role of polyhydroxybutyrate and glycogen as carbon storage compounds in pea and bean bacteroids
Rhizobium leguminosarum synthesizes polyhydroxybutyrate and glycogen as its main carbon storage compounds. To examine the role of these compounds in bacteroid development and in symbiotic efficiency, single and double mutants of R. leguminosarum bv. viciae were made which lack polyhydroxybutyrate synthase (phaC), glycogen synthase (glgA), or both. For comparison, a single phaC mutant also was isolated in a bean-nodulating strain of R. leguminosarum bv. phaseoli. In one large glasshouse trial, the growth of pea plants inoculated with the R. leguminosarum bv. viciae phaC mutant were significantly reduced compared with wild-type-inoculated plants. However, in subsequent glasshouse and growth-room studies, the growth of pea plants inoculated with the mutant were similar to wildtype-inoculated plants. Bean plants were unaffected by the loss of polyhydroxybutyrate biosynthesis in bacteroids. Pea plants nodulated by a glycogen synthase mutant, or the glgA/phaC double mutant, grew as well as the wild type in growth-room experiments. Light and electron micrographs revealed that pea nodules infected with the glgA mutant accumulated large amounts of starch in the II/III interzone. This suggests that glycogen may be the dominant carbon storage compound in pea bacteroids. Polyhydroxybutyrate was present in bacteria in the infection thread of pea plants but was broken down during bacteroid formation. In nodules infected with a phaC mutant of R. leguminosarum bv. viciae, there was a drop in the amount of starch in the II/III interzone, where bacteroids form. Therefore, we propose a carbon burst hypothesis for bacteroid formation, where polyhydroxybutyrate accumulated by bacteria is degraded to fuel bacteroid differentiation.
Grafting between model legumes demonstrates roles for roots and shoots in determining nodule type and host/rhizobia specificity
Previous grafting experiments have demonstrated that legume shoots play a critical role in symbiotic development of nitrogen-fixing root nodules by regulating nodule number. Here, reciprocal grafting experiments between the model legumes Lotus japonicus and Medicago truncatula were carried out to investigate the role of the shoot in the host-specificity of legume-rhizobia symbiosis and nodule type. Lotus japonicus is nodulated by Mesorhizobium loti and makes determinate nodules, whereas M. truncatula is nodulated by Sinorhizobium meliloti and makes indeterminate nodules. When inoculated with M. loti, L. japonicus roots grafted on M. truncatula shoots produced determinate nodules identical in appearance to those produced on L. japonicus self-grafted roots. Moreover, the hypernodulation phenotype of L. japonicus har1-1 roots grafted on wild-type M. truncatula shoots was restored to wild type when nodulated with M. loti. Thus, L. japonicus shoots appeared to be interchangeable with M. truncatula shoots in the L. japonicus root/M. loti symbiosis. However, M. truncatula roots grafted on L. japonicus shoots failed to induce nodules after inoculation with S. meliloti or a mixture of S. meliloti and M. loti. Instead, only early responses to S. meliloti such as root hair tip swelling and deformation, plus induction of the early nodulation reporter gene MtENOD11:GUS were observed. The results indicate that the L. japonicus shoot does not support normal symbiosis between the M. truncatula root and its microsymbiont S. meliloti, suggesting that an unidentified shoot-derived factor may be required for symbiotic progression in indeterminate nodules.
Integrative plant biology: role of phloem long-distance macromolecular trafficking
Long-distance control of nodulation: molecules and models
Legume plants develop root nodules to recruit nitrogen-fixing bacteria called rhizobia. This symbiotic relationship allows the host plants to grow even under nitrogen limiting environment. Since nodule development is an energetically expensive process, the number of nodules should be tightly controlled by the host plants. For this purpose, legume plants utilize a long-distance signaling known as autoregulation of nodulation (AON). AON signaling in legumes has been extensively studied over decades but the underlying molecular mechanism had been largely unclear until recently. With the advent of the model legumes, L. japonicus and M. truncatula, we have been seeing a great progress including isolation of the AON-associated receptor kinase. Here, we summarize recent studies on AON and discuss an updated view of the long-distance control of nodulation.
TOO MUCH LOVE, a root regulator associated with the long-distance control of nodulation in Lotus japonicus
Legume plants tightly control the development and number of symbiotic root nodules. In Lotus japonicus, this regulation requires HAR1 (a CLAVATA1-like receptor kinase) in the shoots, suggesting that a long-distance communication between the shoots and the roots may exist. To better understand its molecular basis, we isolated and characterized a novel hypernodulating mutant of L. japonicus named too much love (tml). Compared with the wild type, tml mutants produced much more nodules which densely covered a wider range of the roots. Reciprocal grafting showed that tml hypernodulation is determined by the root genotype. Moreover, grafting a har1 shoot onto a tml rootstock did not exhibit any obvious additive effects on the nodule number, which was further supported by double mutational analysis. These observations indicate that a shoot factor HAR1 and a root factor TML participate in the same genetic pathway which governs the long-distance signaling of nodule number control. We also showed that the inhibitory effect of TML on nodulation is likely to be local. Therefore, TML may function downstream of HAR1 and the gene product TML might serve as a receptor or mediator of unknown mobile signal molecules that are transported from the shoots to the roots.
Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive CCaMK
A novel family in Medicago truncatula consisting of more than 300 nodule- specific genes coding for small, secreted polypeptides with conserved cysteine motifs
An ERF transcription factor in Medicago truncatula that is essential for nod factor signal transduction
Rhizobial bacteria activate the formation of nodules on the appropriate host legume plant, and this requires the bacterial signaling molecule Nod factor. Perception of Nod factor in the plant leads to the activation of a number of rhizobial-induced genes. Putative transcriptional regulators in the GRAS family are known to function in Nod factor signaling, but these proteins have not been shown to be capable of direct DNA binding. Here, we identify an ERF transcription factor, ERF Required for Nodulation (ERN), which contains a highly conserved AP2 DNA binding domain, that is necessary for nodulation. Mutations in this gene block the initiation and development of rhizobial invasion structures, termed infection threads, and thus block nodule invasion by the bacteria. We show that ERN is necessary for Nod factor-induced gene expression and for spontaneous nodulation activated by the calcium- and calmodulin-dependent protein kinase, DMI3, which is a component of the Nod factor signaling pathway. We propose that ERN is a component of the Nod factor signal transduction pathway and functions downstream of DMI3 to activate nodulation gene expression.
A Ca2+/calmodulin- dependent protein kinase required for symbiotic nodule development, gene identification by transcript-based cloning
In the establishment of the legume-rhizobial symbiosis, bacterial lipochitooligosaccharide signaling molecules termed Nod factors activate the formation of a novel root organ, the nodule. Nod factors elicit several responses in plant root hair cells, including oscillations in cytoplasmic calcium levels (termed calcium spiking) and alterations in root hair growth. A number of plant mutants with defects in the Nod factor signaling pathway have been identified. One such Medicago truncatula mutant, dmi3, exhibits calcium spiking and root hair swelling in response to Nod factor, but fails to initiate symbiotic gene expression or cell divisions for nodule formation. On the basis of these data, it is thought that the dmi3 mutant perceives Nod factor but fails to transduce the signal downstream of calcium spiking. Additionally, the dmi3 mutant is defective in the symbiosis with mycorrhizal fungi, indicating the importance of the encoded protein in multiple symbioses. We report the identification of the DMI3 gene, using a gene cloning method based on transcript abundance. We show that transcript-based cloning is a valid approach for cloning genes in barley, indicating the value of this technology in crop plants. DMI3 encodes a calcium/calmodulin-dependent protein kinase. Mutants in pea sym9 have phenotypes similar to dmi3 and have alterations in this gene. The DMI3 class of proteins is well conserved among plants that interact with mycorrhizal fungi, but it is less conserved in Arabidopsis thaliana, which does not participate in the mycorrhizal symbiosis.
The involvement of a cysteine proteinase in the nodule development in Chinese milk vetch infected with Mesorhizobium huakuii subsp. rengei
Developmental biology of a plant-prokaryote symbiosis, the legume root nodule
The development of nitrogen fixing root nodules on the roots of leguminous plants is induced by soil bacteria (for example, from the genus Rhizobium). The formation of this plant organ involves specific activation of genes in both plant and bacterium. Analysis of these genes gives insight into the way in which plant and bacterium succeed in coordinating plant development.
HAR1 mediates systemic regulation of symbiotic organ development
Symbiotic root nodules are beneficial to leguminous host plants; however, excessive nodulation damages the host because it interferes with the distribution of nutrients in the plant. To keep a steady balance, the nodulation programme is regulated systemically in leguminous hosts. Leguminous mutants that have lost this ability display a hypernodulating phenotype. Through the use of reciprocal and self-grafting studies using Lotus japonicus hypernodulating mutants, har1 (also known as sym78), we show that the shoot genotype is responsible for the negative regulation of nodule development. A map-based cloning strategy revealed that HAR1 encodes a protein with a relative molecular mass of 108,000, which contains 21 leucine-rich repeats, a single transmembrane domain and serine/threonine kinase domains. The har1 mutant phenotype was rescued by transfection of the HAR1 gene. In a comparison of Arabidopsis receptor-like kinases, HAR1 showed the highest level of similarity with CLAVATA1 (CLV1). CLV1 negatively regulates formation of the shoot and floral meristems through cell-cell communication involving the CLV3 peptide. Identification of hypernodulation genes thus indicates that genes in leguminous plants bearing a close resemblance to CLV1 regulate nodule development systemically, by means of organ-organ communication.
A Lotus basic leucine zipper protein with a RING-finger motif negatively regulates the developmental program of nodulation
The developmental program of nodulation is regulated systemically in leguminous host species. A mutant astray (Ljsym77) in Lotus japonicus has lost some sort of its ability to regulate this symtem, and shows enhanced and early nodulation. In the absence of rhizobia, this mutant exhibits characteristics associated with defects in light and gravity responses. These nonsymbiotic phenotypes of astray are very similar to those observed in photomorphogenic Arabidopsis mutant hy5. Based on this evidence, we predicted that astray might contain a mutation in the HY5 homologue of L. japonicus. The homologue, named LjBzf, encodes a basic leucine zipper protein in the C-terminal half that shows the highest level of identity with HY5 of all Arabidopsis proteins. It also encodes legume-characteristic combination of motifs, including a RING-finger motif and an acidic region in the N-terminal half. The astray phenotypes were cosegregated with LjBzf, and the failure to splice the intron was detected. Nonsymbiotic and symbiotic phenotypes of astray were complemented by introduction of CaMV35SLjBzf. It is noteworthy that although Arabidopsis hy5 showed an enhancement of lateral root initiation, Lotus astray showed an enhancement of nodule initiation but not of lateral root initiation. Legume-characteristic combination of motifs of ASTRAY may play specific roles in the regulation of nodule development.
Arabidopsis CLV3 peptide directly binds CLV1 ectodomain
CLV1, which encodes a leucine-rich repeat receptor kinase, and CLV3, which encodes a secreted peptide, function in the same genetic pathway to maintain stem cell populations in Arabidopsis shoot apical meristem. Here, we show biochemical evidence, by ligand binding assay and photoaffinity labeling, that the CLV3 peptide directly binds the CLV1 ectodomain with a dissociation constant of 17.5 nM. The CLV1 ectodomain also interacts with the structurally related CLE peptides, with distinct affinities depending on the specific amino acid sequence. Our results provide direct evidence that CLV3 and CLV1 function as a ligand-receptor pair involved in stem cell maintenance.
klavier (klv), a novel hypernodulation mutant of Lotus japonicus affected in vascular tissue organization and floral induction
Nod factor/nitrate- induced CLE genes that drive HAR1-mediated systemic regulation of nodulation
Host legumes control root nodule numbers by sensing external and internal cues. A major external cue is soil nitrate, whereas a feedback regulatory system in which earlier formed nodules suppress further nodulation through shoot-root communication is an important internal cue. The latter is known as autoregulation of nodulation (AUT), and is believed to consist of two long-distance signals: a root-derived signal that is generated in infected roots and transmitted to the shoot; and a shoot-derived signal that systemically inhibits nodulation. In Lotus japonicus, the leucine-rich repeat receptor-like kinase, HYPERNODULATION ABERRANT ROOT FORMATION 1 (HAR1), mediates AUT and nitrate inhibition of nodulation, and is hypothesized to recognize the root-derived signal. Here we identify L. japonicus CLE-Root Signal 1 (LjCLE-RS1) and LjCLE-RS2 as strong candidates for the root-derived signal. A hairy root transformation study shows that overexpressing LjCLE-RS1 and -RS2 inhibits nodulation systemically and, furthermore, that the systemic suppression depends on HAR1. Moreover, LjCLE-RS2 expression is strongly up-regulated in roots by nitrate addition. Based on these findings, we propose a simple model for AUT and nitrate inhibition of nodulation mediated by LjCLE-RS1, -RS2 peptides and the HAR1 receptor-like kinase.
Evidence for structurally specific negative feedback in the Nod factor signal transduction pathway
The Medicago truncatula DMI1 protein modulates cytosolic calcium signaling
In addition to establishing symbiotic relationships with arbuscular mycorrhizal fungi, legumes also enter into a nitrogen-fixing symbiosis with rhizobial bacteria that results in the formation of root nodules. Several genes involved in the development of both arbuscular mycorrhiza and legume nodulation have been cloned in model legumes. Among them, Medicago truncatula DMI1 (DOESN'T MAKE INFECTIONS1) is required for the generation of nucleus-associated calcium spikes in response to the rhizobial signaling molecule Nod factor. DMI1 encodes a membrane protein with striking similarities to the Methanobacterium thermoautotrophicum potassium channel (MthK). The cytosolic C terminus of DMI1 contains a RCK (regulator of the conductance of K(+)) domain that in MthK acts as a calcium-regulated gating ring controlling the activity of the channel. Here we show that a dmi1 mutant lacking the entire C terminus acts as a dominant-negative allele interfering with the formation of nitrogen-fixing nodules and abolishing the induction of calcium spikes by the G-protein agonist Mastoparan. Using both the full-length DMI1 and this dominant-negative mutant protein we show that DMI1 increases the sensitivity of a sodium- and lithium-hypersensitive yeast (Saccharomyces cerevisiae) mutant toward those ions and that the C-terminal domain plays a central role in regulating this response. We also show that DMI1 greatly reduces the release of calcium from internal stores in yeast, while the dominant-negative allele appears to have the opposite effect. This work suggests that DMI1 is not directly responsible for Nod factor-induced calcium changes, but does have the capacity to regulate calcium channels in both yeast and plants.
The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations
Metabolic changes of rhizobia in legume nodules
Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases
Although most higher plants establish a symbiosis with arbuscular mycorrhizal fungi, symbiotic nitrogen fixation with rhizobia is a salient feature of legumes. Despite this host range difference, mycorrhizal and rhizobial invasion shares a common plant-specified genetic programme controlling the early host interaction. One feature distinguishing legumes is their ability to perceive rhizobial-specific signal molecules. We describe here two LysM-type serine/threonine receptor kinase genes, NFR1 and NFR5, enabling the model legume Lotus japonicus to recognize its bacterial microsymbiont Mesorhizobium loti. The extracellular domains of the two transmembrane kinases resemble LysM domains of peptidoglycan- and chitin-binding proteins, suggesting that they may be involved directly in perception of the rhizobial lipochitin-oligosaccharide signal. We show that NFR1 and NFR5 are required for the earliest physiological and cellular responses to this lipochitin-oligosaccharide signal, and demonstrate their role in the mechanism establishing susceptibility of the legume root for bacterial infection.
The symbiotic ion channel homolog DMI1 is localized in the nuclear membrane of Medicago truncatula roots
A plant regulator controlling development of symbiotic root nodules
Symbiotic nitrogen-fixing root nodules on legumes are founded by root cortical cells that de-differentiate and restart cell division to establish nodule primordia. Bacterial microsymbionts invade these primordia through infection threads laid down by the plant and, after endocytosis, membrane-enclosed bacteroids occupy cells in the nitrogen-fixing tissue of functional nodules. The bacteria excrete lipochitin oligosaccharides, triggering a developmental process that is controlled by the plant and can be suppressed. Nodule inception initially relies on cell competence in a narrow infection zone located just behind the growing root tip. Older nodules then regulate the number of nodules on a root system by suppressing the development of nodule primordia. To identify the regulatory components that act early in nodule induction, we characterized a transposon-tagged Lotus japonicus mutant, nin (for nodule inception), arrested at the stage of bacterial recognition. We show that nin is required for the formation of infection threads and the initiation of primordia. NIN protein has regional similarity to transcription factors, and the predicted DNA-binding/dimerization domain identifies and typifies a consensus motif conserved in plant proteins with a function in nitrogen-controlled development.
The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length
Four Medicago truncatula sunn mutants displayed shortened roots and hypernodulation under all conditions examined. The mutants, recovered in three independent genetic screens, all contained lesions in a leucine-rich repeat (LRR) receptor kinase. Although the molecular defects among alleles varied, root length and the extent of nodulation were not significantly different between the mutants. SUNN is expressed in shoots, flowers and roots. Although previously reported grafting experiments showed that the presence of the mutated SUNN gene in roots does not confer an obvious phenotype, expression levels of SUNN mRNA were reduced in sunn-1 roots. SUNN and the previously identified genes HAR1 (Lotus japonicus) and NARK (Glycine max) are orthologs based on gene sequence and synteny between flanking sequences. Comparison of related LRR receptor kinases determined that all nodulation autoregulation genes identified to date are the closest legume relatives of AtCLV1 by sequence, yet sunn, har and nark mutants do not display the fasciated clv phenotype. The M. truncatula region is syntenic with duplicated regions of Arabidopsis chromosomes 2 and 4, none of which harbor CLV1 or any other LRR receptor kinase genes. A novel truncated copy of the SUNN gene lacking a kinase domain, RLP1, is found immediately upstream of SUNN and like SUNN is expressed at a reduced level in sunn-1 roots.
Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase
Proliferation of legume nodule primordia is controlled by shoot-root signaling known as autoregulation of nodulation (AON). Mutants defective in AON show supernodulation and increased numbers of lateral roots. Here, we demonstrate that AON in soybean is controlled by the receptor-like protein kinase GmNARK (Glycine max nodule autoregulation receptor kinase), similar to Arabidopsis CLAVATA1 (CLV1). Whereas CLV1 functions in a protein complex controlling stem cell proliferation by short-distance signaling in shoot apices, GmNARK expression in the leaf has a major role in long-distance communication with nodule and lateral root primordia.
Nod factor elicits two separable calcium responses in Medicago truncatula root hair cells
Modulation of intracellular calcium levels plays a key role in the transduction of many biological signals. Here, we characterize early calcium responses of wild-type and mutant Medicago truncatula plants to nodulation factors produced by the bacterial symbiont Sinorhizobium meliloti using a dual-dye ratiometric imaging technique. When presented with 1 nM Nod factor, root hair cells exhibited only the previously described calcium spiking response initiating 10 min after application. Nod factor (10 nM) elicited an immediate increase in calcium levels that was temporally earlier and spatially distinct from calcium spikes occurring later in the same cell. Nod factor analogs that were structurally related, applied at 10 nM, failed to initiate this calcium flux response. Cells induced to spike with low Nod factor concentrations show a calcium flux response when Nod factor is raised from 1 to 10 nM. Plant mutants previously shown to be deficient for the calcium spiking response (dmi1 and dmi2) exhibited an immediate, truncated calcium flux with 10 nM Nod factor, demonstrating a competence to respond to Nod factor but an impaired ability to generate a full biphasic response. These results demonstrate that the legume root hair cell exhibits two independent calcium responses to Nod factor triggered at different agonist concentrations and suggests an early branch point in the Nod factor signal transduction pathway.
Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling
Rhizobia secrete nodulation (Nod) factors, which set in motion the formation of nitrogen-fixing root nodules on legume host plants. Nod factors induce several cellular responses in root hair cells within minutes, but also are essential for the formation of infection threads by which rhizobia enter the root. Based on studies using bacterial mutants, a two-receptor model was proposed, a signaling receptor that induces early responses with low requirements toward Nod factor structure and an entry receptor that controls infection with more stringent demands. Recently, putative Nod factor receptors were shown to be LysM domain receptor kinases. However, mutants in these receptors, in both Lotus japonicus (nfr1 and nfr5) and Medicago truncatula (Medicago; nfp), do not support the two-receptor model because they lack all Nod factor-induced responses. LYK3, the putative Medicago ortholog of NFR1, has only been studied by RNA interference, showing a role in infection thread formation. Medicago hair curling (hcl) mutants are unable to form curled root hairs, a step preceding infection thread formation. We identified the weak hcl-4 allele that is blocked during infection thread growth. We show that HCL encodes LYK3 and, thus, that this receptor, besides infection, also controls root hair curling. By using rhizobial mutants, we also show that HCL controls infection thread formation in a Nod factor structure-dependent manner. Therefore, LYK3 functions as the proposed entry receptor, specifically controlling infection. Finally, we show that LYK3, which regulates a subset of Nod factor-induced genes, is not required for the induction of NODULE INCEPTION.
NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription
Rhizobial Nod factors induce in their legume hosts the expression of many genes and set in motion developmental processes leading to root nodule formation. Here we report the identification of the Medicago GRAS-type protein Nodulation signaling pathway 1 (NSP1), which is essential for all known Nod factor-induced changes in gene expression. NSP1 is constitutively expressed, and so it acts as a primary transcriptional regulator mediating all known Nod factor-induced transcriptional responses, and therefore, we named it a Nod factor response factor.
Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression
The Rhizobium-legume symbiosis culminates in the exchange of nutrients in the root nodule. Bacteria within the nodule reduce molecular nitrogen for plant use and plants provide bacteria with carbon-containing compounds. Following the initial signaling events that lead to plant infection, little is known about the plant requirements for establishment and maintenance of the symbiosis. We screened 44,000 M2 plants from fast neutron-irradiated Medicago truncatula seeds and isolated eight independent mutant lines that are defective in nitrogen fixation. The eight mutants are monogenic and represent seven complementation groups. To monitor bacterial status in mutant nodules, we assayed Sinorhizobium meliloti symbiosis gene promoters (nodF, exoY, bacA, and nifH) in the defective in nitrogen fixation mutants. Additionally, we used an Affymetrix oligonucleotide microarray to monitor gene expression changes in wild-type and three mutant plants during the nodulation process. These analyses suggest the mutants can be separated into three classes: one class that supports little to no nitrogen fixation and minimal bacterial expression of nifH; another class that supports no nitrogen fixation and minimal bacterial expression of nodF, bacA, and nifH; and a final class that supports low levels of both nitrogen fixation and bacterial nifH expression.
A plant receptor-like kinase required for both bacterial and fungal symbiosis
Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum
Symbiosis, inventiveness by recruitment?
crinkle, a novel symbiotic mutant that affects the infection thread growth and alters the root hair, trichome, and seed development in Lotus japonicus
To elucidate the mechanisms involved in Rhizobium-legume symbiosis, we examined a novel symbiotic mutant, crinkle (Ljsym79), from the model legume Lotus japonicus. On nitrogen-starved medium, crinkle mutants inoculated with the symbiont bacterium Mesorhizobium loti MAFF 303099 showed severe nitrogen deficiency symptoms. This mutant was characterized by the production of many bumps and small, white, uninfected nodule-like structures. Few nodules were pale-pink and irregularly shaped with nitrogen-fixing bacteroids and expressing leghemoglobin mRNA. Morphological analysis of infected roots showed that nodulation in crinkle mutants is blocked at the stage of the infection process. Confocal microscopy and histological examination of crinkle nodules revealed that infection threads were arrested upon penetrating the epidermal cells. Starch accumulation in uninfected cells and undeveloped vascular bundles were also noted in crinkle nodules. Results suggest that the Crinkle gene controls the infection process that is crucial during the early stage of nodule organogenesis. Aside from the symbiotic phenotypes, crinkle mutants also developed morphological alterations, such as crinkly or wavy trichomes, short seedpods with aborted embryos, and swollen root hairs. crinkle is therefore required for symbiotic nodule development and for other aspects of plant development.
api, a novel Medicago truncatula symbiotic mutant impaired in nodule primordium invasion
Genetic approaches have proved to be extremely useful in dissecting the complex nitrogen-fixing Rhizobium-legume endosymbiotic association. Here we describe a novel Medicago truncatula mutant called api, whose primary phenotype is the blockage of rhizobial infection just prior to nodule primordium invasion, leading to the formation of large infection pockets within the cortex of noninvaded root outgrowths. The mutant api originally was identified as a double symbiotic mutant associated with a new allele (nip-3) of the NIP/LATD gene, following the screening of an ethylmethane sulphonate-mutagenized population. Detailed characterization of the segregating single api mutant showed that rhizobial infection is also defective at the earlier stage of infection thread (IT) initiation in root hairs, as well as later during IT growth in the small percentage of nodules which overcome the primordium invasion block. Neither modulating ethylene biosynthesis (with L-alpha-(2-aminoethoxyvinylglycine or 1-aminocyclopropane-1-carboxylic acid) nor reducing ethylene sensitivity in a skl genetic background alters the basic api phenotype, suggesting that API function is not closely linked to ethylene metabolism or signaling. Genetic mapping places the API gene on the upper arm of the M. truncatula linkage group 4, and epistasis analyses show that API functions downstream of BIT1/ERN1 and LIN and upstream of NIP/LATD and the DNF genes.
Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements
In situ immunolocalization of tubulin revealed that important rearrangements occur during all the early symbiotic steps in the Medicago/R. meliloti symbiotic interaction. Microtubular cytoskeleton (MtC) reorganizations were observed in inner tissues, first in the pericycle and then in the inner cortex where the nodule primordium forms. Subsequently, major MtC changes occurred in outer tissues, associated with root hair activation and curling, the formation of preinfection threads (PITs) and the initiation and the growth of an infection network. From the observed sequence of MtC changes, we propose a model which aims to better define, at the histological level, the timing of the early symbiotic stages. This model suggests the existence of two opposite gradients of cell differentiation controlling respectively the formation of division centers in the inner cortex and plant preparation for infection. It implies that (i) MtC rearrangements occur in pericycle and inner cortex earlier than in the root hair, (ii) the infection process proceeds prior to the formation of the nodule meristem, (iii) the initial primordium prefigures the future zone II of the mature nodule and (iv) the nodule meristem derives from the nodule primordium. Finally, our data also strongly suggest that in alfalfa PIT differentiation, a stage essential for successful infection, requires complementary signaling additional to Nod factors.
Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development
Induced development of a new plant organ in response to rhizobia is the most prominent manifestation of legume root-nodule symbiosis with nitrogen-fixing bacteria. Here we show that the complex root-nodule organogenic programme can be genetically deregulated to trigger de novo nodule formation in the absence of rhizobia or exogenous rhizobial signals. In an ethylmethane sulphonate-induced snf1 (spontaneous nodule formation) mutant of Lotus japonicus, a single amino-acid replacement in a Ca2+/calmodulin-dependent protein kinase (CCaMK) is sufficient to turn fully differentiated root cortical cells into meristematic founder cells of root nodule primordia. These spontaneous nodules are genuine nodules with an ontogeny similar to that of rhizobial-induced root nodules, corroborating previous physiological studies. Using two receptor-deficient genetic backgrounds we provide evidence for a developmentally integrated spontaneous nodulation process that is independent of lipochitin-oligosaccharide signal perception and oscillations in Ca2+ second messenger levels. Our results reveal a key regulatory position of CCaMK upstream of all components required for cell-cycle activation, and a phenotypically divergent series of mutant alleles demonstrates positive and negative regulation of the process.
Induction of pre-infection thread structures in the leguminous host plant by mitogenic Lipooligosaccharides of Rhizobium
Root nodules of leguminous plants are symbiotic organs in which Rhizobium bacteria fix nitrogen. Their formation requires the induction of a nodule meristem and the formation of a tubular structure, the infection thread, through which the rhizobia reach the nodule primordium. In the Rhizobium host plants pea and vetch, pre-infection thread structures always preceded the formation of infection threads. These structures consisted of cytoplasmic bridges traversing the central vacuole of outer cortical root cells, aligned in radial rows. In vetch, the site of the infection thread was determined by the plant rather than by the invading rhizobia. Like nodule primordia, pre-infection thread structures could be induced in the absence of rhizobia provided that mitogenic lipo-oligosaccharides produced by Rhizobium leguminosarum biovar viciae were added to the plant. In this case, cells in the two outer cortical cell layers containing cytoplasmic bridges may have formed root hairs. A common morphogenetic pathway may be shared in the formation of root hairs and infection threads.
Suggested nomenclature for plant genes involved in nodulation and symbiosis
Cell wall degradation during infection thread formation by the root nodule bacterium Rhizobium leguminosarum is a two-step process
In the nitrogen-fixing root-nodule symbiosis of Rhizobium leguminosarum biovar viciae and its host plants pea and vetch, the bacteria enter one root cortical cell after another via a tip-growing structure, the infection thread. Rhizobial Nod (nodulation) factors induce the formation of preinfection thread structures (Van Brussel, A.A.N., R. Bakhuizen, P.C. van Spronsen, H.P. Spaink, T. Tak, B.J.J. Lugtenberg, J.W. Kijne, Science 257, 70-72 (1992)), but formation of infection threads requires the presence of bacterial cells. Passing of an infection thread from cell to cell requires local cell wall degradation. We compared at the ultrastructural level local cell wall changes in the outer root cortex of pea and vetch related to preinfection thread formation and infection thread formation, respectively. Cell wall modifications in the outer periclinal walls of root cortical cells induced by Nod factors appeared to be similar to those induced by rhizobia. These modifications take place opposite cytoplasmic bridges and are probably related to induction of tip growth. However, complete cell wall degradation was never observed in the absence of rhizobia. We propose a two-step cell wall degradation process for infection thread formation. The first step is a local cell wall modification by plant enzymes, induced by rhizobial Nod factors. The second step is complete cell wall degradation in the presence of rhizobia.
Cell biological changes of outer cortical root cells in early determinate nodulation
In the symbiosis of leguminous plants and Rhizobium bacteria, nodule primordia develop in the root cortex. This can be either in the inner cortex (indeterminate-type of nodulation) or outer cortex (determinate-type of nodulation), depending upon the host plant. We studied and compared early nodulation stages in common bean (Phaseolus vulgaris) and Lotus japonicus, both known as determinate-type nodulation plants. Special attention was paid to the occurrence of cytoplasmic bridges, the influence of rhizobial Nod factors (lipochitin oligosaccharides [LCOs]) on this phenomenon, and sensitivity of the nodulation process to ethylene. Our results show that i) both plant species form initially broad, matrix-rich infection threads; ii) cytoplasmic bridges occur in L. japonicus but not in bean; iii) formation of these bridges is induced by rhizobial LCOs; iv) formation of primordia starts in L. japonicus in the middle root cortex and in bean in the outer root cortex; and v) in the presence of the ethylene-biosynthesis inhibitor aminoethoxyvinylglycine (AVG), nodulation of L. japonicus is stimulated when the roots are grown in the light, which is consistent with the role of cytoplasmic bridges during nodulation of L. japonicus.
Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules
Bacteroid differentiation was examined in developing and mature alfalfa nodules elicited by wild-type or Fix- mutant strains of Rhizobium meliloti. Ultrastructural studies of wild-type nodules distinguished five steps in bacteroid differentiation (types 1 to 5), each being restricted to a well-defined histological region of the nodule. Correlative studies between nodule development, bacteroid differentiation, and acetylene reduction showed that nitrogenase activity was always associated with the differentiation of the distal zone III of the nodule. In this region, the invaded cells were filled with heterogeneous type 4 bacteroids, the cytoplasm of which displayed an alternation of areas enriched with ribosomes or with DNA fibrils. Cytological studies of complementary halves of transversally sectioned mature nodules confirmed that type 4 bacteroids were always observed in the half of the nodule expressing nitrogenase activity, while the presence of type 5 bacteroids could never be correlated with acetylene reduction. Bacteria with a transposon Tn5 insertion in pSym fix genes elicited the development of Fix- nodules in which bacteroids could not develop into the last two ultrastructural types. The use of mutant strains deleted of DNA fragments bearing functional reiterated pSym fix genes and complemented with recombinant plasmids, each carrying one of these fragments, strengthened the correlation between the occurrence of type 4 bacteroids and acetylene reduction. A new nomenclature is proposed to distinguish the histological areas in alfalfa nodules which account for and are correlated with the multiple stages of bacteroid development.
nip, a symbiotic Medicago truncatula mutant that forms root nodules with aberrant infection threads and plant defense-like response
To investigate the legume-Rhizobium symbiosis, we isolated and studied a novel symbiotic mutant of the model legume Medicago truncatula, designated nip (numerous infections and polyphenolics). When grown on nitrogen-free media in the presence of the compatible bacterium Sinorhizobium meliloti, the nip mutant showed nitrogen deficiency symptoms. The mutant failed to form pink nitrogen-fixing nodules that occur in the wild-type symbiosis, but instead developed small bump-like nodules on its roots that were blocked at an early stage of development. Examination of the nip nodules by light microscopy after staining with X-Gal for S. meliloti expressing a constitutive GUS gene, by confocal microscopy following staining with SYTO-13, and by electron microscopy revealed that nip initiated symbiotic interactions and formed nodule primordia and infection threads. The infection threads in nip proliferated abnormally and very rarely deposited rhizobia into plant host cells; rhizobia failed to differentiate further in these cases. nip nodules contained autofluorescent cells and accumulated a brown pigment. Histochemical staining of nip nodules revealed this pigment to be polyphenolic accumulation. RNA blot analyses demonstrated that nip nodules expressed only a subset of genes associated with nodule organogenesis, as well as elevated expression of a host defense-associated phenylalanine ammonia lyase gene. nip plants were observed to have abnormal lateral roots. nip plant root growth and nodulation responded normally to ethylene inhibitors and precursors. Allelism tests showed that nip complements 14 other M. truncatula nodulation mutants but not latd, a mutant with a more severe nodulation phenotype as well as primary and lateral root defects. Thus, the nip mutant defines a new locus, NIP, required for appropriate infection thread development during invasion of the nascent nodule by rhizobia, normal lateral root elongation, and normal regulation of host defense-like responses during symbiotic interactions.
EFD is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula
Mechanisms regulating legume root nodule development are still poorly understood, and very few regulatory genes have been cloned and characterized. Here, we describe EFD (for ethylene response factor required for nodule differentiation), a gene that is upregulated during nodulation in Medicago truncatula. The EFD transcription factor belongs to the ethylene response factor (ERF) group V, which contains ERN1, 2, and 3, three ERFs involved in Nod factor signaling. The role of EFD in the regulation of nodulation was examined through the characterization of a null deletion mutant (efd-1), RNA interference, and overexpression studies. These studies revealed that EFD is a negative regulator of root nodulation and infection by Rhizobium and that EFD is required for the formation of functional nitrogen-fixing nodules. EFD appears to be involved in the plant and bacteroid differentiation processes taking place beneath the nodule meristem. We also showed that EFD activated Mt RR4, a cytokinin primary response gene that encodes a type-A response regulator. We propose that EFD induction of Mt RR4 leads to the inhibition of cytokinin signaling, with two consequences: the suppression of new nodule initiation and the activation of differentiation as cells leave the nodule meristem. Our work thus reveals a key regulator linking early and late stages of nodulation and suggests that the regulation of the cytokinin pathway is important both for nodule initiation and development.
Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula
The symbiotic interaction between Medicago truncatula and Sinorhizobium meliloti results in the formation of nitrogen-fixing nodules on the roots of the host plant. The early stages of nodule formation are induced by bacteria via lipochitooligosaccharide signals known as Nod factors (NFs). These NFs are structurally specific for bacterium-host pairs and are sufficient to cause a range of early responses involved in the host developmental program. Early events in the signal transduction of NFs are not well defined. We have previously reported that Medicago sativa root hairs exposed to NF display sharp oscillations of cytoplasmic calcium ion concentration (calcium spiking). To assess the possible role of calcium spiking in the nodulation response, we analyzed M. truncatula mutants in five complementation groups. Each of the plant mutants is completely Nod- and is blocked at early stages of the symbiosis. We defined two genes, DMI1 and DMI2, required in common for early steps of infection and nodulation and for calcium spiking. Another mutant, altered in the DMI3 gene, has a similar mutant phenotype to dmi1 and dmi2 mutants but displays normal calcium spiking. The calcium behavior thus implies that the DMI3 gene acts either downstream of calcium spiking or downstream of a common branch point for the calcium response and the later nodulation responses. Two additional mutants, altered in the NSP and HCL genes, which show root hair branching in response to NF, are normal for calcium spiking. This system provides an opportunity to use genetics to study ligand-stimulated calcium spiking as a signal transduction event.
Structure-function analysis of Nod factor-induced root hair calcium spiking in Rhizobium-legume symbiosis
In the Rhizobium-legume symbiosis, compatible bacteria and host plants interact through an exchange of signals: Host compounds promote the expression of bacterial biosynthetic nod (nodulation) genes leading to the production of a lipochito-oligosaccharide signal, the Nod factor (NF). The particular array of nod genes carried by a given species of Rhizobium determines the NF structure synthesized and defines the range of legume hosts by which the bacterium is recognized. Purified NF can induce early host responses even in the absence of live Rhizobium One of the earliest known host responses to NF is an oscillatory behavior of cytoplasmic calcium, or calcium spiking, in root hair cells, initially observed in Medicago spp. and subsequently characterized in four other genera (D.W. Ehrhardt, R. Wais, S.R. Long [1996] Cell 85: 673-681; S.A. Walker, V. Viprey, J.A. Downie [2000] Proc Natl Acad Sci USA 97: 13413-13418; D.W. Ehrhardt, J.A. Downie, J. Harris, R.J. Wais, and S.R. Long, unpublished data). We sought to determine whether live Rhizobium trigger a rapid calcium spiking response and whether this response is NF dependent. We show that, in the Sinorhizobium meliloti-Medicago truncatula interaction, bacteria elicit a calcium spiking response that is indistinguishable from the response to purified NF. We determine that calcium spiking is a nod gene-dependent host response. Studies of calcium spiking in M. truncatula and alfalfa (Medicago sativa) also uncovered the possibility of differences in early NF signal transduction. We further demonstrate the sufficiency of the nod genes for inducing calcium spiking by using Escherichia coli BL21 (DE3) engineered to express 11 S. meliloti nod genes.
Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by Nod factors and chitin oligomers
Changes in intracellular calcium in pea root hairs responding to Rhizobium leguminosarum bv. viciae nodulation (Nod) factors were analyzed by using a microinjected calcium-sensitive fluorescent dye (dextran-linked Oregon Green). Within 1-2 min after Nod-factor addition, there was usually an increase in fluorescence, followed about 10 min later by spikes in fluorescence occurring at a rate of about one spike per minute. These spikes, corresponding to an increase in calcium of approximately 200 nM, were localized around the nuclear region, and they were similar in terms of lag and period to those induced by Nod factors in alfalfa. Calcium responses were analyzed in nonnodulating pea mutants, representing seven loci that affect early stages of the symbiosis. Mutations affecting three loci (sym8, sym10, and sym19) abolished Nod-factor-induced calcium spiking, whereas a normal response was seen in peas carrying alleles of sym2(A), sym7, sym9, and sym30. Chitin oligomers of four or five N-acetylglucosamine residues could also induce calcium spiking, although the response was qualitatively different from that induced by Nod factors; a rapid increase in intracellular calcium was not observed, the period between spikes was lower, and the response was not as sustained. The chitin-oligomer-induced calcium spiking did not occur in nodulation mutants (sym8, sym10, and sym19) that were defective for Nod-factor-induced spiking, suggesting that this response is related to nodulation signaling. From our data and previous observations on the lack of mycorrhizal infection in some of the sym mutants, we propose a model for the potential order of pea nodulation genes in nodulation and mycorrhizal signaling.
Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia
Legumes form symbioses with rhizobia, which initiate the development of a new plant organ, the nodule. Flavonoids have long been hypothesized to regulate nodule development through their action as auxin transport inhibitors, but genetic proof has been missing. To test this hypothesis, we used RNA interference to silence chalcone synthase (CHS), the enzyme that catalyzes the first committed step of the flavonoid pathway, in Medicago truncatula. Agrobacterium rhizogenes transformation was used to create hairy roots that showed strongly reduced CHS transcript levels and reduced levels of flavonoids in silenced roots. Flavonoid-deficient roots were unable to initiate nodules, even though normal root hair curling was observed. Nodule formation and flavonoid accumulation could be rescued by supplementation of plants with the precursor flavonoids naringenin and liquiritigenin. The flavonoid-deficient roots showed increased auxin transport compared with control roots. Inoculation with rhizobia reduced auxin transport in control roots after 24 h, similar to the action of the auxin transport inhibitor N-(1-naphthyl)phthalamic acid (NPA). Rhizobia were unable to reduce auxin transport in flavonoid-deficient roots, even though NPA inhibited auxin transport. Our results present genetic evidence that root flavonoids are necessary for nodule initiation in M. truncatula and suggest that they act as auxin transport regulators.