

Chin J Plant Ecol ›› 2025, Vol. 49 ›› Issue (2): 308-319.DOI: 10.17521/cjpe.2023.0220 cstr: 32100.14.cjpe.2023.0220
• Research Articles • Previous Articles Next Articles
YIN Si1, YANG Yi-Ting1, LU Rui-Ling1, NIAN Rui1, HAO Zhuan2,*( ), GAO Yong1,*(
), GAO Yong1,*( )
)
Received:2023-08-01
Accepted:2023-12-21
Online:2025-02-20
Published:2025-02-20
Contact:
HAO Zhuan, GAO Yong
Supported by:YIN Si, YANG Yi-Ting, LU Rui-Ling, NIAN Rui, HAO Zhuan, GAO Yong. Phylogeographic study of natural populations of Amorphophallus yunnanensis (Araceae) in China[J]. Chin J Plant Ecol, 2025, 49(2): 308-319.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.plant-ecology.com/EN/10.17521/cjpe.2023.0220
| 编号 No. | 种群编号 Population ID | 纬度 Latitude (N) | 经度 Longitude (E) | 海拔 Altitude (m) | 采样点 Sample Site |
|---|---|---|---|---|---|
| 1 | YSXS | 28.78° | 110.22° | 317 | 湖南永顺 Yongshun, Hunan |
| 2 | LDNR | 25.40° | 106.61° | 436 | 贵州罗甸 Luodian, Guizhou |
| 3 | WMNS | 25.16° | 106.25° | 726 | 贵州望谟 Wangmo, Guizhou |
| 4 | WMXY | 25.22° | 106.12° | 1 075 | 贵州望谟 Wangmo, Guizhou |
| 5 | WMZX | 24.98° | 106.12° | 432 | 贵州望谟 Wangmo, Guizhou |
| 6 | LYLZ | 24.28° | 106.67° | 396 | 广西凌云 Lingyun, Guangxi |
| 7 | FHG | 24.61° | 104.25° | 1 249 | 云南师宗 Shizong, Yunnan |
| 8 | SQG | 24.96° | 102.63° | 2 216 | 云南昆明 Kunming, Yunnan |
| 9 | FYND | 24.25° | 102.19° | 1 527 | 云南玉溪 Yuxi, Yunnan |
| 10 | BMGD | 22.66° | 101.16° | 1 680 | 云南普洱 Pu’er, Yunnan |
| 11 | LCYD | 24.24° | 99.58° | 1 329 | 云南临沧 Lincang, Yunnan |
| 12 | HHLC | 23.02° | 102.38° | 1 715 | 云南红河 Honghe, Yunnan |
| 13 | HDR | 21.95° | 100.41° | 1 177 | 云南西双版纳 Xishuangbanna, Yunnan |
| 14 | DSQ | 24.73° | 100.51° | 2 340 | 云南大理 Dali, Yunnan |
| 15 | JDXC | 24.26° | 101.05° | 1 754 | 云南景东 Jingdong, Yunnan |
| 16 | ZYHC | 23.96° | 100.96° | 1 897 | 云南镇沅 Zhenyuan, Yunnan |
Table 1 Geographic information of natural populations of Amorphophallus yunnanensis in China
| 编号 No. | 种群编号 Population ID | 纬度 Latitude (N) | 经度 Longitude (E) | 海拔 Altitude (m) | 采样点 Sample Site |
|---|---|---|---|---|---|
| 1 | YSXS | 28.78° | 110.22° | 317 | 湖南永顺 Yongshun, Hunan |
| 2 | LDNR | 25.40° | 106.61° | 436 | 贵州罗甸 Luodian, Guizhou |
| 3 | WMNS | 25.16° | 106.25° | 726 | 贵州望谟 Wangmo, Guizhou |
| 4 | WMXY | 25.22° | 106.12° | 1 075 | 贵州望谟 Wangmo, Guizhou |
| 5 | WMZX | 24.98° | 106.12° | 432 | 贵州望谟 Wangmo, Guizhou |
| 6 | LYLZ | 24.28° | 106.67° | 396 | 广西凌云 Lingyun, Guangxi |
| 7 | FHG | 24.61° | 104.25° | 1 249 | 云南师宗 Shizong, Yunnan |
| 8 | SQG | 24.96° | 102.63° | 2 216 | 云南昆明 Kunming, Yunnan |
| 9 | FYND | 24.25° | 102.19° | 1 527 | 云南玉溪 Yuxi, Yunnan |
| 10 | BMGD | 22.66° | 101.16° | 1 680 | 云南普洱 Pu’er, Yunnan |
| 11 | LCYD | 24.24° | 99.58° | 1 329 | 云南临沧 Lincang, Yunnan |
| 12 | HHLC | 23.02° | 102.38° | 1 715 | 云南红河 Honghe, Yunnan |
| 13 | HDR | 21.95° | 100.41° | 1 177 | 云南西双版纳 Xishuangbanna, Yunnan |
| 14 | DSQ | 24.73° | 100.51° | 2 340 | 云南大理 Dali, Yunnan |
| 15 | JDXC | 24.26° | 101.05° | 1 754 | 云南景东 Jingdong, Yunnan |
| 16 | ZYHC | 23.96° | 100.96° | 1 897 | 云南镇沅 Zhenyuan, Yunnan |
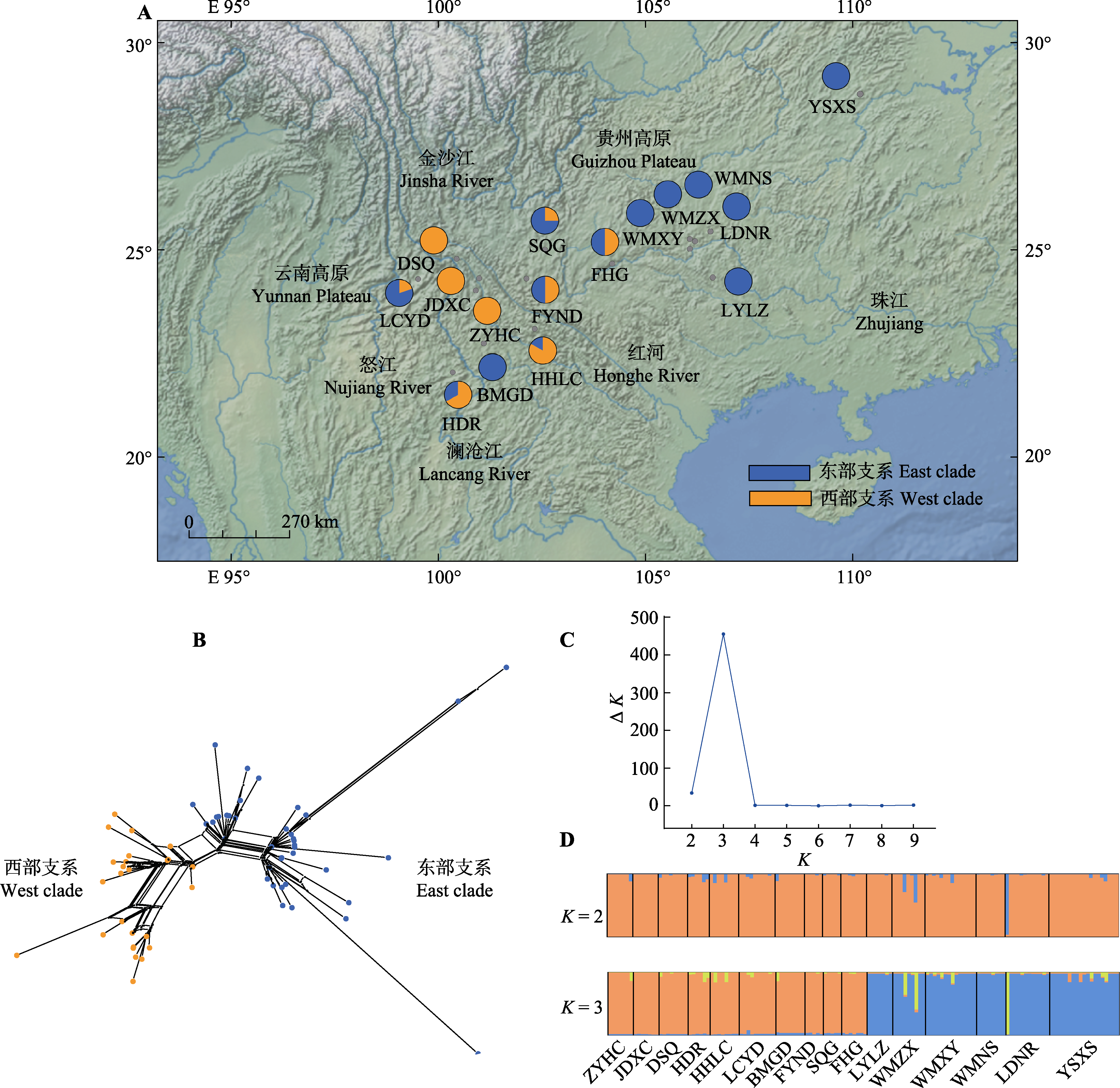
Fig. 1 Geographic distributions of cpDNA haplotypes found in 16 sampled populations of Amorphophallus yunnanensis in China. A, Geographic locations of 16 sampled populations and distributions of cpDNA haplotypes within two phylogenetic branches. B, Neighbor-net haplotype tree for Amorphophallus yunnanensis based on cpDNA haplotypes. C, ΔK values of the posterior probability distribution estimated by STRUCTURE HARVESTER. D, Bar plot of population assign proportions to each genetic cluster at K = 2 and K = 3. K, the number of genetic clusters. Population ID see Table 1.
| 标记名称 Marker ID | 正向引物 Forward Primer (5′-3′) | 反向引物 Reverse Primer (5′-3′) | 退火温度 Annealing temperature (℃) |
|---|---|---|---|
| trnK-matK | CTTGCAGTTTTCATTGCACA | TTCACTTTTGGTCTCAACCC | 56 |
| rbcL | ATGTCACAACAAACAGAAAC | TCCTTTTAGTAAAAGATTGGGCCGAG | 56 |
| trnL | CGAAATCGGTAGACGCTACG | GGGGATAGAGGGACTTGAAC | 56 |
| psaB | AAATATCACAAGTACCACCTCG | ACAATCGGATTACGCACCA | 56 |
| rps2 | CTGGAATCGAAATATCTGC | GTATCAACGGTCAATCCTC | 56 |
Table 2 Sequences of cpDNA primers used in phylogeographic study Amorphophallus yunnanensis in China
| 标记名称 Marker ID | 正向引物 Forward Primer (5′-3′) | 反向引物 Reverse Primer (5′-3′) | 退火温度 Annealing temperature (℃) |
|---|---|---|---|
| trnK-matK | CTTGCAGTTTTCATTGCACA | TTCACTTTTGGTCTCAACCC | 56 |
| rbcL | ATGTCACAACAAACAGAAAC | TCCTTTTAGTAAAAGATTGGGCCGAG | 56 |
| trnL | CGAAATCGGTAGACGCTACG | GGGGATAGAGGGACTTGAAC | 56 |
| psaB | AAATATCACAAGTACCACCTCG | ACAATCGGATTACGCACCA | 56 |
| rps2 | CTGGAATCGAAATATCTGC | GTATCAACGGTCAATCCTC | 56 |
| 单倍型群组 Haplogroup | 种群编号 Population ID | 样本数 n | 单倍型 Haplotype | 单倍型多样性 Hd | 突变位点 S | 核苷酸多样性 π | Tajima’s D | Fu’s Fs |
|---|---|---|---|---|---|---|---|---|
| 东部支系 East clade | 104 | 0.826 | 87 | 0.000 94 | -31.168* | -2.567* | ||
| YSXS | 19 | H_17, 64, 65, 66, 67, 68, 69 | 0.538 | 9 | 0.000 29 | -1.999* | -2.533* | |
| LDNR | 12 | H_17, 20, 21, 22, 23, 50 | 0.818 | 40 | 0.001 82 | 1.968* | -2.153* | |
| WMNS | 8 | H_17, 18, 19, 20 | 0.643 | 6 | 0.000 39 | -0.422 | -1.640 | |
| WMXY | 14 | H_16, 17, 24, 25, 26, 27, 28, 29, 30 | 0.835 | 15 | 0.000 72 | -3.436* | -1.836* | |
| WMZX | 9 | H_17, 58, 59, 60 | 0.643 | 16 | 0.001 55 | 2.661 | -0.172 | |
| LYLZ | 7 | H_17, 51, 52 | 0.524 | 1 | 0.000 07 | -0.095 | -1.006 | |
| FHG | 7 | H_12, 53, 54, 55, 56, 57 | 0.857 | 5 | 0.000 54 | -1.447 | 0.132 | |
| SQG | 5 | H_12, 61, 62, 63 | 0.900 | 3 | 0.000 31 | -1.938 | -1.048 | |
| FYND | 5 | H_12, 13, 14, 15 | 0.900 | 5 | 0.000 62 | -0.701 | 0.000 | |
| BMGD | 8 | H_1, 2 | 0.250 | 4 | 0.000 26 | -1.535 | 1.946 | |
| LCYD | 10 | H_45, 46, 47, 48, 49 | 0.756 | 10 | 0.000 59 | -0.318 | -1.590 | |
| 西部支系 West clade | 36 | 0.887 | 27 | 0.000 96 | -12.337* | -1.503 | ||
| HHLC | 8 | H_31, 32, 33, 34, 35, 36 | 0.929 | 12 | 0.000 78 | -1.609 | -0.665 | |
| HDR | 6 | H_3, 4, 5, 6, 37, 38 | 1.000 | 11 | 0.001 07 | -2.552 | -0.859 | |
| DSQ | 8 | H_7, 8, 9, 10, 11 | 0.786 | 5 | 0.000 32 | -2.238 | -1.595 | |
| JDXC | 7 | H_9, 39, 40, 41, 42, 43, 44 | 1.000 | 4 | 0.000 42 | -0.538 | -0.040 | |
| ZYHC | 7 | H_9, 11, 70, 71 | 0.714 | 2 | 0.000 20 | -0.438 | -0.275 |
Table 3 Genetic diversity of the 16 populations of Amorphophallus yunnanensis in China based on cpDNA data
| 单倍型群组 Haplogroup | 种群编号 Population ID | 样本数 n | 单倍型 Haplotype | 单倍型多样性 Hd | 突变位点 S | 核苷酸多样性 π | Tajima’s D | Fu’s Fs |
|---|---|---|---|---|---|---|---|---|
| 东部支系 East clade | 104 | 0.826 | 87 | 0.000 94 | -31.168* | -2.567* | ||
| YSXS | 19 | H_17, 64, 65, 66, 67, 68, 69 | 0.538 | 9 | 0.000 29 | -1.999* | -2.533* | |
| LDNR | 12 | H_17, 20, 21, 22, 23, 50 | 0.818 | 40 | 0.001 82 | 1.968* | -2.153* | |
| WMNS | 8 | H_17, 18, 19, 20 | 0.643 | 6 | 0.000 39 | -0.422 | -1.640 | |
| WMXY | 14 | H_16, 17, 24, 25, 26, 27, 28, 29, 30 | 0.835 | 15 | 0.000 72 | -3.436* | -1.836* | |
| WMZX | 9 | H_17, 58, 59, 60 | 0.643 | 16 | 0.001 55 | 2.661 | -0.172 | |
| LYLZ | 7 | H_17, 51, 52 | 0.524 | 1 | 0.000 07 | -0.095 | -1.006 | |
| FHG | 7 | H_12, 53, 54, 55, 56, 57 | 0.857 | 5 | 0.000 54 | -1.447 | 0.132 | |
| SQG | 5 | H_12, 61, 62, 63 | 0.900 | 3 | 0.000 31 | -1.938 | -1.048 | |
| FYND | 5 | H_12, 13, 14, 15 | 0.900 | 5 | 0.000 62 | -0.701 | 0.000 | |
| BMGD | 8 | H_1, 2 | 0.250 | 4 | 0.000 26 | -1.535 | 1.946 | |
| LCYD | 10 | H_45, 46, 47, 48, 49 | 0.756 | 10 | 0.000 59 | -0.318 | -1.590 | |
| 西部支系 West clade | 36 | 0.887 | 27 | 0.000 96 | -12.337* | -1.503 | ||
| HHLC | 8 | H_31, 32, 33, 34, 35, 36 | 0.929 | 12 | 0.000 78 | -1.609 | -0.665 | |
| HDR | 6 | H_3, 4, 5, 6, 37, 38 | 1.000 | 11 | 0.001 07 | -2.552 | -0.859 | |
| DSQ | 8 | H_7, 8, 9, 10, 11 | 0.786 | 5 | 0.000 32 | -2.238 | -1.595 | |
| JDXC | 7 | H_9, 39, 40, 41, 42, 43, 44 | 1.000 | 4 | 0.000 42 | -0.538 | -0.040 | |
| ZYHC | 7 | H_9, 11, 70, 71 | 0.714 | 2 | 0.000 20 | -0.438 | -0.275 |
| 种群编号 Population ID | BMGD | FYND | WMNS | LDNR | WMXY | LCYD | LYLZ | FHG | WMZX | SQG | YSXS | HDR | DSQ | HHLC | JDXC | ZYHC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BMGD | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |
| FYND | 0.386 | * | * | * | * | * | NS | * | NS | * | * | * | * | * | * | |
| WMNS | 0.686 | 0.517 | NS | NS | * | NS | * | NS | * | NS | * | * | * | * | * | |
| LDNR | 0.379 | 0.232 | -0.011 | * | * | NS | * | NS | * | * | * | * | * | * | * | |
| WMXY | 0.556 | 0.385 | 0.034 | 0.027 | * | NS | * | * | * | * | * | * | * | * | * | |
| LCYD | 0.446 | 0.216 | 0.538 | 0.346 | 0.481 | * | * | * | * | * | * | * | * | * | * | |
| LYLZ | 0.762 | 0.584 | -0.008 | -0.025 | 0.038 | 0.565 | * | NS | * | NS | * | * | * | * | * | |
| FHG | 0.381 | -0.085 | 0.509 | 0.273 | 0.396 | 0.258 | 0.560 | * | NS | * | * | * | * | * | * | |
| WMZX | 0.466 | 0.321 | 0.071 | 0.053 | 0.115 | 0.423 | 0.062 | 0.349 | * | * | * | * | * | * | * | |
| SQG | 0.448 | 0.018 | 0.588 | 0.233 | 0.409 | 0.246 | 0.711 | 0.005 | 0.323 | * | * | * | * | * | * | |
| YSXS | 0.641 | 0.506 | 0.008 | 0.046 | 0.062 | 0.563 | -0.022 | 0.507 | 0.135 | 0.531 | * | * | * | * | * | |
| HDR | 0.484 | 0.152 | 0.550 | 0.349 | 0.471 | 0.380 | 0.580 | 0.188 | 0.408 | 0.271 | 0.584 | * | * | * | * | |
| DSQ | 0.615 | 0.366 | 0.686 | 0.372 | 0.553 | 0.437 | 0.762 | 0.368 | 0.466 | 0.448 | 0.639 | 0.472 | * | NS | NS | |
| HHLC | 0.465 | 0.143 | 0.538 | 0.353 | 0.464 | 0.369 | 0.563 | 0.180 | 0.419 | 0.253 | 0.571 | 0.277 | 0.452 | * | * | |
| JDXC | 0.472 | 0.159 | 0.552 | 0.334 | 0.468 | 0.354 | 0.588 | 0.188 | 0.406 | 0.249 | 0.574 | 0.299 | 0.042 | 0.298 | NS | |
| ZYHC | 0.514 | 0.281 | 0.595 | 0.334 | 0.506 | 0.375 | 0.646 | 0.310 | 0.419 | 0.318 | 0.599 | 0.411 | -0.005 | 0.385 | 0.034 |
Table 4 Genetic differentiation (pairwise FST) among populations of Amorphophallus yunnanensis based on cpDNA data
| 种群编号 Population ID | BMGD | FYND | WMNS | LDNR | WMXY | LCYD | LYLZ | FHG | WMZX | SQG | YSXS | HDR | DSQ | HHLC | JDXC | ZYHC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BMGD | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |
| FYND | 0.386 | * | * | * | * | * | NS | * | NS | * | * | * | * | * | * | |
| WMNS | 0.686 | 0.517 | NS | NS | * | NS | * | NS | * | NS | * | * | * | * | * | |
| LDNR | 0.379 | 0.232 | -0.011 | * | * | NS | * | NS | * | * | * | * | * | * | * | |
| WMXY | 0.556 | 0.385 | 0.034 | 0.027 | * | NS | * | * | * | * | * | * | * | * | * | |
| LCYD | 0.446 | 0.216 | 0.538 | 0.346 | 0.481 | * | * | * | * | * | * | * | * | * | * | |
| LYLZ | 0.762 | 0.584 | -0.008 | -0.025 | 0.038 | 0.565 | * | NS | * | NS | * | * | * | * | * | |
| FHG | 0.381 | -0.085 | 0.509 | 0.273 | 0.396 | 0.258 | 0.560 | * | NS | * | * | * | * | * | * | |
| WMZX | 0.466 | 0.321 | 0.071 | 0.053 | 0.115 | 0.423 | 0.062 | 0.349 | * | * | * | * | * | * | * | |
| SQG | 0.448 | 0.018 | 0.588 | 0.233 | 0.409 | 0.246 | 0.711 | 0.005 | 0.323 | * | * | * | * | * | * | |
| YSXS | 0.641 | 0.506 | 0.008 | 0.046 | 0.062 | 0.563 | -0.022 | 0.507 | 0.135 | 0.531 | * | * | * | * | * | |
| HDR | 0.484 | 0.152 | 0.550 | 0.349 | 0.471 | 0.380 | 0.580 | 0.188 | 0.408 | 0.271 | 0.584 | * | * | * | * | |
| DSQ | 0.615 | 0.366 | 0.686 | 0.372 | 0.553 | 0.437 | 0.762 | 0.368 | 0.466 | 0.448 | 0.639 | 0.472 | * | NS | NS | |
| HHLC | 0.465 | 0.143 | 0.538 | 0.353 | 0.464 | 0.369 | 0.563 | 0.180 | 0.419 | 0.253 | 0.571 | 0.277 | 0.452 | * | * | |
| JDXC | 0.472 | 0.159 | 0.552 | 0.334 | 0.468 | 0.354 | 0.588 | 0.188 | 0.406 | 0.249 | 0.574 | 0.299 | 0.042 | 0.298 | NS | |
| ZYHC | 0.514 | 0.281 | 0.595 | 0.334 | 0.506 | 0.375 | 0.646 | 0.310 | 0.419 | 0.318 | 0.599 | 0.411 | -0.005 | 0.385 | 0.034 |

Fig. 2 Phylogenetic inference for the relationships between haplotypes of Amorphophallus yunnanensis based on Bayesian analysis. Posterior probabilities, divergence times, and confidence intervals are shown at each node. Scale bar indicates five substitution per site. Population ID see Table 1.

Fig. 3 Median-joining network for the cpDNA haplotypes (H) of Amorphophallus yunnanensis in Southwestern China. The size of each circle is proportional to the individual number of the haplotype.
| 方差来源 Source of variation | 自由度 df | 平方和 Sum of squares | 方差组分 Variance components | 方差比例 Percentage of variation (%) | F-statistics | p |
|---|---|---|---|---|---|---|
| 支系间 Between two clades | 1 | 39.526 | 0.594 22 | 20.24 | FCT = 0.202 | <0.001 |
| 种群间 Among populations | 14 | 112.735 | 0.743 17 | 25.32 | FSC = 0.317 | <0.001 |
| 种群内 Within populations | 124 | 198.181 | 1.598 24 | 54.44 | FST = 0.456 | <0.001 |
Table 5 Analysis of molecular variance (AMOVA) of Amorphophallus yunnanensis populations based on cpDNA sequences
| 方差来源 Source of variation | 自由度 df | 平方和 Sum of squares | 方差组分 Variance components | 方差比例 Percentage of variation (%) | F-statistics | p |
|---|---|---|---|---|---|---|
| 支系间 Between two clades | 1 | 39.526 | 0.594 22 | 20.24 | FCT = 0.202 | <0.001 |
| 种群间 Among populations | 14 | 112.735 | 0.743 17 | 25.32 | FSC = 0.317 | <0.001 |
| 种群内 Within populations | 124 | 198.181 | 1.598 24 | 54.44 | FST = 0.456 | <0.001 |

Fig. 6 Geographic patterns in the habitat suitability of Amorphophallus yunnanensis during the last interglacial (LIG, A), the last glacial maximum (LGM, B), the mid-Holocene (MH, C), and at present (D) evaluated using MaxEnt.
| [1] | Bai LW, Niu Y, Liu HL, Zhang SL (2016). The germplasms and breeding progress of Amorphophallus. South China Agriculture, 10(4), 48-52. |
| [白立伟, 牛义, 刘海利, 张盛林 (2016). 魔芋种质资源及育种研究进展. 南方农业, 10(4), 48-52.] | |
| [2] | Chen SY, Wang JX (2017). Progress in research on tectonic uplift in Yunnan-Guizhou Plateau. Yunnan Geographic Environment Research, 29(3), 23-29. |
| [陈思宇, 王嘉学 (2017). 云贵高原隆升研究进展. 云南地理环境研究, 29(3), 23-29.] | |
| [3] | Deng J, Fu R, Compton SG, Liu M, Wang Q, Yuan C, Zhang L, Chen Y (2020). Sky islands as foci for divergence of fig trees and their pollinators in southwest China. Molecular Ecology, 29, 762-782. |
| [4] |
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution, 29, 1969-1973.
DOI PMID |
| [5] |
Evanno G, Regnaut S, Goudet J (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14, 2611-2620.
DOI PMID |
| [6] |
Excoffier L, Lischer HEL (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567.
DOI PMID |
| [7] | Gao Y (2022). Development of SSR markers for molecular identification of Amorphophallus species. Jounal of Qujing Normal University, 41(3), 46-52. |
| [高永 (2022). 魔芋种间分子鉴定的SSR标记开发. 曲靖师范学院学报, 41(3), 46-52.] | |
| [8] | Gao Y, Yin S, Yang HX, Wu LF, Yan YH (2018). Genetic diversity and phylogenetic relationships of seven Amorphophallus species in southwestern China revealed by chloroplast DNA sequences. Mitochondrial DNA. Part A, DNA Mapping, Sequencing, and Analysis, 29, 679-686. |
| [9] | Gong X, Xie SF, Xia ZR, Wang AX, Tong LT (2023). Study on the effects of adding konjac refined flour on the quality of fresh wet rice noodle. Food & Machinery, 39(7), 186-192. |
| [龚雪, 解松峰, 夏曾润, 王爱霞, 佟立涛 (2023). 魔芋精粉对鲜湿米粉品质的影响. 食品与机械, 39(7), 186-192.] | |
| [10] | Han F, Lamichhaney S, Grant BR, Grant PR, Andersson L, Webster MT (2017). Gene flow, ancient polymorphism, and ecological adaptation shape the genomic landscape of divergence among Darwin’s finches. Genome Research, 27, 1004-1015. |
| [11] | He K, Jiang XL (2014). Sky Islands of southwest China. I: an overview of phylogeographic patterns. Chinese Science Bulletin, 59, 585-597. |
| [12] |
Ho SYW, Shapiro B (2011). Skyline-plot methods for estimating demographic history from nucleotide sequences. Molecular Ecology Resources, 11, 423-434.
DOI PMID |
| [13] | Hu HY, Yang YZ, Li A, Zheng ZY, Zhang J, Liu JQ (2022). Genomic divergence of Stellera chamaejasme through local selection across the Qinghai-Tibet Plateau and Northern China. Molecular Ecology, 31, 4782-4796. |
| [14] |
Huson DH (1998). SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics, 14, 68-73.
PMID |
| [15] |
Hutchison DW, Templeton AR (1999). Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution, 53, 1898-1914.
DOI PMID |
| [16] | Ju MM, Fu Y, Zhao GF, He CZ, Li ZH, Tian B (2018). Effects of the Tanaka Line on the genetic structure of Bombax ceiba (Malvaceae) in dry-hot valley areas of southwest China. Ecology and Evolution, 8, 3599-3608. |
| [17] |
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods, 14, 587-589.
DOI PMID |
| [18] |
Kumar S, Stecher G, Tamura K (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870-1874.
DOI PMID |
| [19] | Li H, Hetterscheid WLA (2010). Amorphophallus//Editorial Committee of Flora of China. Flora of China: Vol. 36. Science Press, Beijing. 23-33. |
| [20] | Li H, Long CL (1998). Taxonomy of Amorphophallus in China. Acta Botanica Yunnanica, 20(2), 167-170. |
| [李恒, 龙春林 (1998). 中国磨芋属的分类问题. 云南植物研究, 20(2), 167-170.] | |
| [21] |
Librado P, Rozas J (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451-1452.
DOI PMID |
| [22] | Liu W, Xie J, Zhou H, Kong H, Hao G, Fritsch PW, Gong W (2021). Population dynamics linked to glacial cycles in Cercis chuniana F. P. Metcalf (Fabaceae) endemic to the montane regions of subtropical China. Evolutionary Applications, 14, 2647-2663. |
| [23] | López-Pujol J, Zhang F, Sun H, Ying T, Ge S (2011). Centres of plant endemism in China: places for survival or for speciation? Journal of Biogeography, 38, 1267-1280. |
| [24] | Meng FJ, Liu L, Peng M, Wang ZK, Wang C, Zhao YY (2015). Genetic diversity and population structure analysis in wild strawberry (Fragaria nubicola L.) from Motuo in Tibet Plateau based on simple sequence repeats (SSRs). Biochemical Systematics and Ecology, 63, 113-118. |
| [25] | Moritz C (1994). Defining ‘evolutionarily significant units’ for conservation. Trends in Ecology & Evolution, 9, 373-375. |
| [26] |
Nauheimer L, Metzler D, Renner SS (2012). Global history of the ancient monocot family Araceae inferred with models accounting for past continental positions and previous ranges based on fossils. New Phytologist, 195, 938-950.
DOI PMID |
| [27] | Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32, 268-274. |
| [28] | Niu Y, Zhang SL, Wang ZM, Li C, Sun YH (2005). Research and utilization of konjac (Amorphophallus) resources in China. Journal of Southwest Agricultural University (Natural Science Edition), 27, 634-638. |
| [牛义, 张盛林, 王志敏, 李川, 孙远航 (2005). 中国魔芋资源的研究与利用. 西南农业大学学报(自然科学版), 27, 634-638.] | |
| [29] |
Orsini L, Vanoverbeke J, Swillen I, Mergeay J, de Meester L (2013). Drivers of population genetic differentiation in the wild: isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Molecular Ecology, 22, 5983-5999.
DOI PMID |
| [30] | Pan C (2012). The Development of Microsatellite Loci in Amorphophallus konjac and Genetic Structure in Amorphophallus. PhD dissertation, Wuhan University, Wuhan. |
| [潘程 (2012). 花魔芋微卫星标记开发及魔芋属植物遗传结构研究. 博士学位论文, 武汉大学, 武汉.] | |
| [31] |
Peakall R, Smouse PE (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics, 28, 2537-2539.
PMID |
| [32] | Phillips SJ, Anderson RP, Schapire RE (2006). Maximum entropy modeling of species geographic distributions. Ecological Modelling, 190, 231-259. |
| [33] | Pouchon C, Gauthier J, Pitteloud C, Claudel C, Alvarez N (2023). Phylogenomic study of Amorphophallus (Alismatales; Araceae): When plastid DNA gene sequences help to resolve the backbone subgeneric delineation. Journal of Systematics and Evolution, 61, 64-79. |
| [34] |
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945-959.
DOI PMID |
| [35] |
Prunier R, Holsinger KE (2010). Was it an explosion? Using population genetics to explore the dynamics of a recent radiation within Protea (Proteaceae L.). Molecular Ecology, 19, 3968-3980.
DOI PMID |
| [36] | Ren PY, Pan MQ (2013). Population genetic structure of five Amorphophallus species from the south of Yunnan Province by inter-simple sequences (ISSR) markers. Journal of Wuhan University (Natural Science Edition), 59(1), 99-104. |
| [任盘宇, 潘明清 (2013). 云南南部5种魔芋属植物居群遗传结构的ISSR分析. 武汉大学学报(理学版), 59(1), 99-104.] | |
| [37] |
Simmons MP, Ochoterena H (2000). Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology, 49, 369-381.
PMID |
| [38] | Stokstad E (2020). Mountains and monsoons created Tibetan biodiversity. Science, 369, 493. DOI: 10.1126/science.369.6503.493. |
| [39] | Sun H, Wang XP, Fan DY, Sun OJ (2022). Contrasting vegetation response to climate change between two monsoon regions in Southwest China: the roles of climate condition and vegetation height. Science of the Total Environment, 802, 149643. DOI: 10.1016/j.scitotenv.2021.149643. |
| [40] | Teng CZ, Diao Y, Chang FHS, Xie SQ, Han YH, Hu ZL (2006). ISSR analyses of relative relationships among germplasm resources of Amorphophallus blume from Yunnan Province. Anhui Agricultural Science Bulletin, 12(11), 54-56. |
| [滕彩珠, 刁英, 常福浩森, 谢世清, 韩永华, 胡中立 (2006). 云南魔芋种质资源亲缘关系的ISSR分析. 安徽农学通报, 12(11), 54-56.] | |
| [41] | Wambulwa MC, Luo YH, Zhu GF, Milne R, Wachira FN, Wu ZY, Wang H, Gao LM, Li DZ, Liu J (2022). Determinants of genetic structure in a highly heterogeneous landscape in southwest China. Frontiers in Plant Science, 13, 779989. DOI: 10.3389/fpls.2022.779989. |
| [42] | Wang B, Mao JF, Zhao W, Wang XR (2013). Impact of geography and climate on the genetic differentiation of the subtropical pine Pinus yunnanensis. PLoS ONE, 8, e67345. DOI: 10.1371/journal.pone.0067345. |
| [43] | Wang BS, Mojica JP, Perera N, Lee CR, Lovell JT, Sharma A, Adam C, Lipzen A, Barry K, Rokhsar DS, Schmutz J, Mitchell-Olds T (2019). Ancient polymorphisms contribute to genome-wide variation by long-term balancing selection and divergent sorting in Boechera stricta. Genome Biology, 20, 126. DOI: 10.1186/s13059-019-1729-9. |
| [44] | Yin S, Chu H, Zhang Y, Lu F, Gao Y (2021) Population genetic diversity and genetic structure of Amorphophallus yunnanensis in Southwestern China and its conservation implication. Taiwania, 66, 126-134. |
| [45] | Yin S, Hao Z, Lu FD, Gao Y (2023). Genetic diversity of six Amorphophallus species in Southwest China based on cpDNA sequences. Guihaia, 43, 2042-2051. |
| [殷斯, 郝转, 陆飞东, 高永 (2023). 西南地区六种魔芋属植物基于cpDNA序列的遗传多样性研究. 广西植物, 43, 2042-2051.] | |
| [46] | Zang RG, Dong M, Li JQ, Chen XY, Zeng SJ, Jiang MX, Li ZQ, Huang JH (2016). Conservation and restoration for typical critically endangered wild plants with extremely small population. Acta Ecologica Sinica, 36, 7130-7135. |
| [臧润国, 董鸣, 李俊清, 陈小勇, 曾宋君, 江明喜, 李镇清, 黄继红 (2016). 典型极小种群野生植物保护与恢复技术研究. 生态学报, 36, 7130-7135.] | |
| [47] | Zhang TC, Comes HP, Sun H (2011). Chloroplast phylogeography of Terminalia franchetii (Combretaceae) from the eastern Sino-Himalayan region and its correlation with historical river capture events. Molecular Phylogenetics and Evolution, 60, 1-12. |
| [1] | 黄 承玲, Han Li Rong, Ling Qing Hong, Xiong Yang Sheng, Ling Tian Xiao, Xia Guowei Xia Guowei, Ren Chen Zheng, Wei Zhou. Study on genetic conservation of Rhododendron liboense based on SNP molecular markers,a plant species with extremely small populations [J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [2] | ZHANG Hong-Xiang, WEN Zhi-Bin, WANG Qian. Population genetic structure of Malus sieversii and environmental adaptations [J]. Chin J Plant Ecol, 2022, 46(9): 1098-1108. |
| [3] | WANG Chun-Cheng, MA Song-Mei, ZHANG Dan, WANG Shao-Ming. Spatial genetic structure of Lycium ruthenicum in the Qaidam Basin [J]. Chin J Plant Ecol, 2020, 44(6): 661-668. |
| [4] | CHAI Yong-Fu, XU Jin-Shi, LIU Hong-Yan, LIU Quan-Ru, ZHENG Cheng-Yang, KANG Mu-Yi, LIANG Cun-Zhu, WANG Ren-Qing, GAO Xian-Ming, ZHANG Feng, SHI Fu-Chen, LIU Xiao, YUE Ming. Species composition and phylogenetic structure of major shrublands in North China [J]. Chin J Plant Ecol, 2019, 43(9): 793-805. |
| [5] | Ming-Fei ZHAO, Feng XUE, Yu-Hang WANG, Guo-Yi WANG, Kai-Xiong XING, Mu-Yi KANG, Jing-Lan WANG. Phylogenetic structure and diversity of herbaceous communities in the conifer forests along an elevational gradient in Luya Mountain, Shanxi, China [J]. Chin J Plan Ecolo, 2017, 41(7): 707-715. |
| [6] | HUANG Jian-Xiong, ZHENG Feng-Ying, MI Xiang-Cheng. Influence of environmental factors on phylogenetic structure at multiple spatial scales in an evergreen broad-leaved forest of China [J]. Chin J Plant Ecol, 2010, 34(3): 309-315. |
| [7] | YAN Bang-Guo, WEN Wei-Quan, ZHANG Jian, YANG Wan-Qin, LIU Yang, HUANG Xu, LI Ze-Bo. Plant community assembly rules across a subalpine grazing gradient in western Sichuan, China [J]. Chin J Plant Ecol, 2010, 34(11): 1294-1302. |
| [8] | CHEN Liang-Hua, HU Ting-Xing, ZHANG Fan, LI Guo-He. GENETIC DIVERSITIES OF FOUR JUGLANSPOPULATIONS REVEALED BY AFLP IN SICHUAN PROVINCE, CHINA [J]. Chin J Plant Ecol, 2008, 32(6): 1362-1372. |
| [9] | WANG Ying, KANG Ming, HUANG Hong-Wen. SUBPOPULATION GENETIC STRUCTURE IN A PANMICTIC POPULATION AS REVEALED BY MOLECULAR MARKERS: A CASE STUDY OF CASTANEA SEQUINII USING SSR MARKERS [J]. Chin J Plant Ecol, 2006, 30(1): 147-156. |
| [10] | CHEN Xiao-Yong. Spatial Autocorrelation of Genetic Structure in a Population of Cyclobalanopsis glauca in Huangshan, Anhui [J]. Chin J Plan Ecolo, 2001, 25(1): 29-34. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2026 Chinese Journal of Plant Ecology
Tel: 010-62836134, 62836138, E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
![]()