

Chin J Plant Ecol ›› 2022, Vol. 46 ›› Issue (7): 735-752.DOI: 10.17521/cjpe.2021.0386
• Review • Next Articles
WU Kai1, LI Kai1,*( ), JIA Wei-Han2,3, LIAO Meng-Na1, NI Jian1
), JIA Wei-Han2,3, LIAO Meng-Na1, NI Jian1
Received:2021-10-28
Accepted:2022-01-26
Online:2022-07-20
Published:2022-02-16
Contact:
LI Kai
Supported by:WU Kai, LI Kai, JIA Wei-Han, LIAO Meng-Na, NI Jian. Modern processes of lacustrine plant sedimentary ancient DNA[J]. Chin J Plant Ecol, 2022, 46(7): 735-752.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.plant-ecology.com/EN/10.17521/cjpe.2021.0386
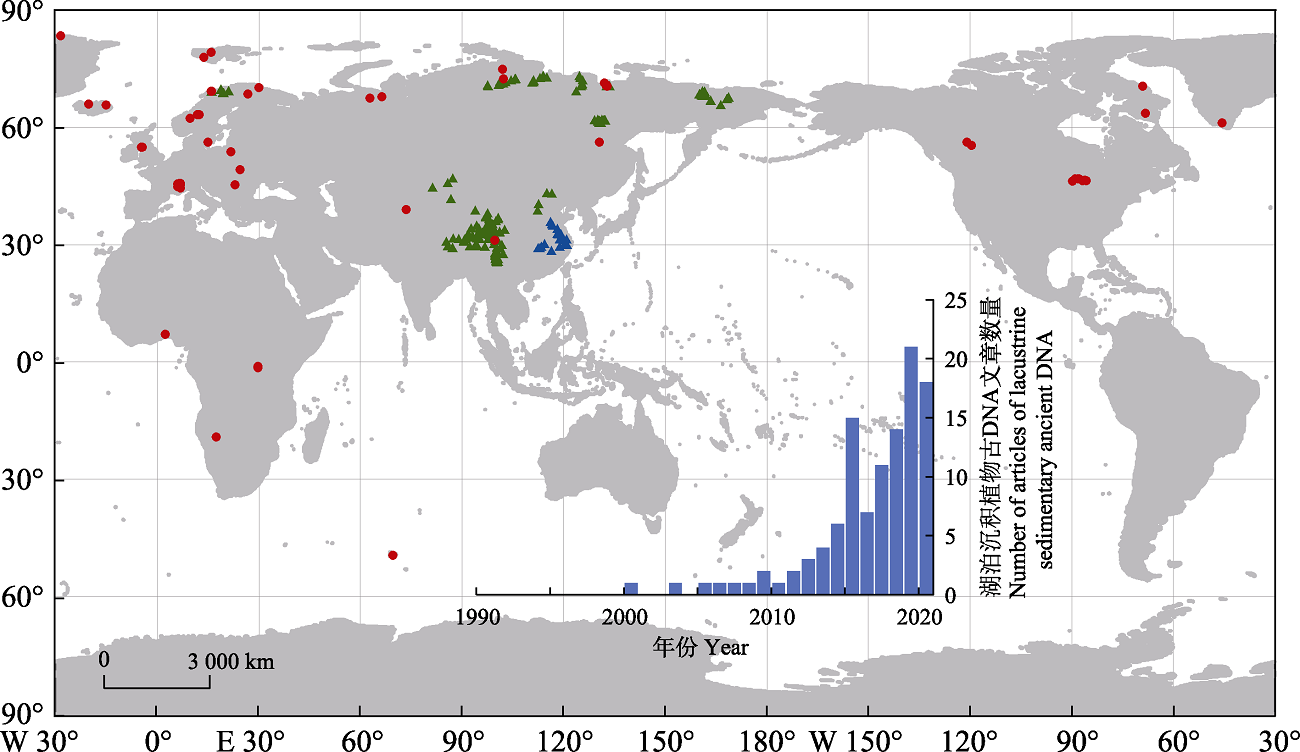
Fig. 1 Study sites of lacustrine plant sedimentary ancient DNA. Red circles represent the core sediment study sites (the references are shown in Table 1), the green triangles represent the lake surface sediment study sites (Niemeyer et al., 2017; Alsos et al., 2018; Jia et al., 2022), and the blue triangles represent the lake surface sedimentary plant DNA study sites being conducted by our research group (unpublished data). The histogram shows the change in the number of publications retrieved from the Web of Science that focus on the sedimentary ancient plant DNA.
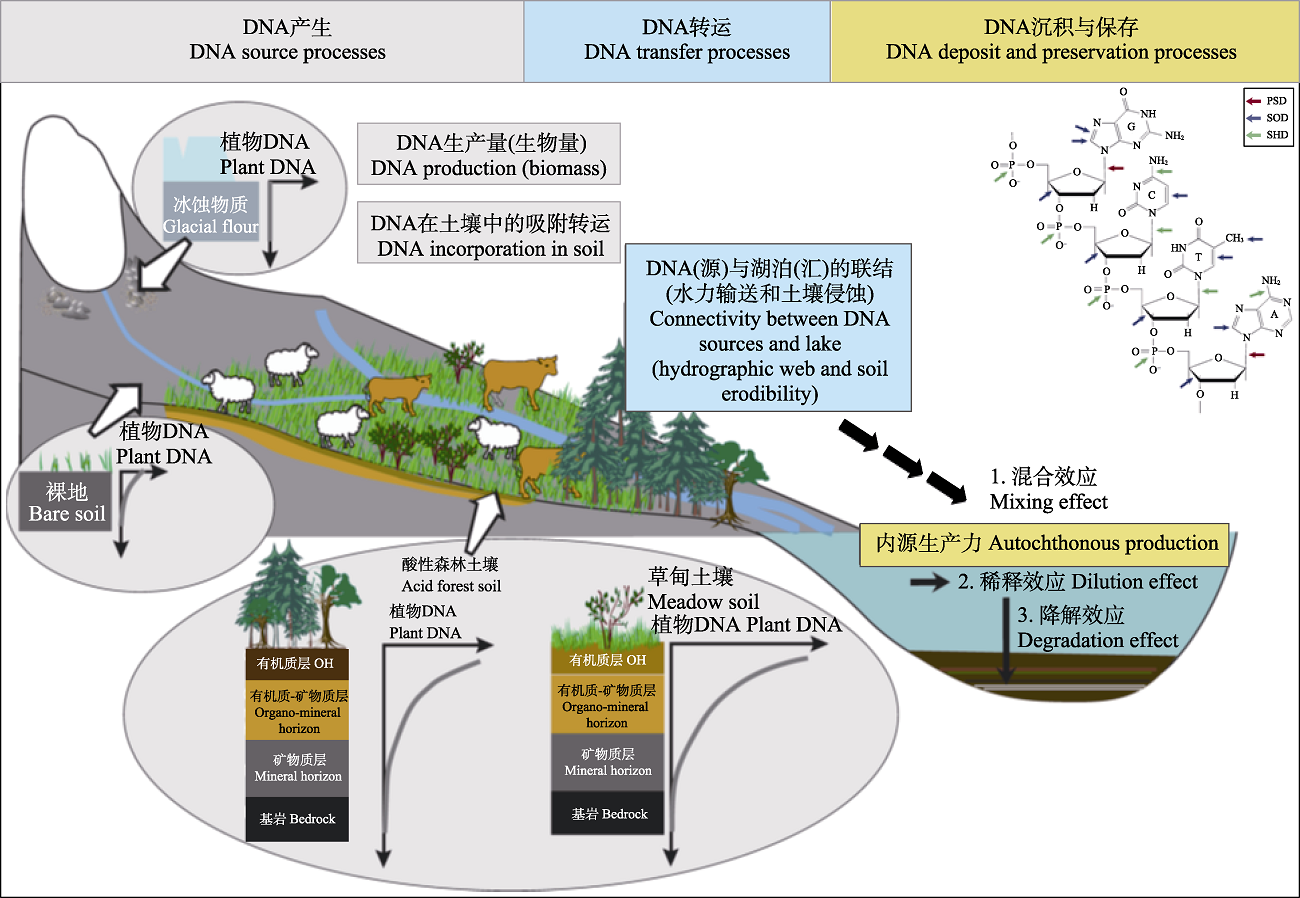
Fig. 2 Process model of production, transportation, deposition, and preservation of plant DNA (tissue DNA and extracellular DNA) at catchment scale (modified from Giguet-Covex et al. (2019)). The upper right corner shows the targeting sites (Lindahl, 1993; Blum et al., 1997; Strickler et al., 2015) where DNA degradation mainly occurs. The degradation process of DNA exists in the whole process, while the particle adsorption of soil and sediment and the preservation of DNA in sediment compensate for the degradation effect of DNA to some extent. The distribution of extracellular plant DNA in soil profiles has a decreasing trend from top to bottom. Acidic soils and bare soils would comprise little extracellular plant DNA. Moreover, glacial flour is free of extracellular plant DNA. PSD, principal site of damage (depurination); SHD, site of hydrolytic damage; SOD, site of oxidative damage. OH, organic horizon (litter + humus).
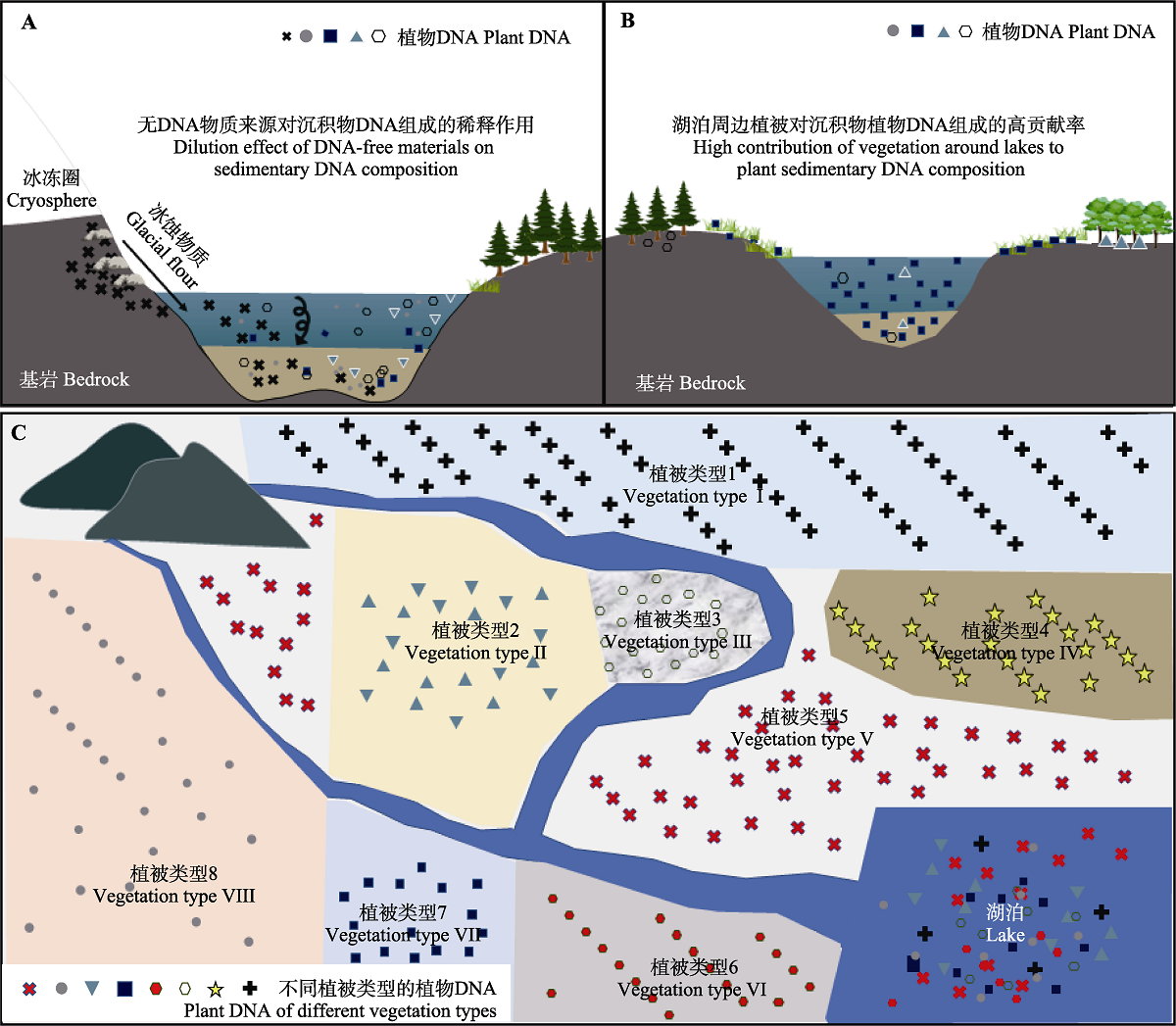
Fig. 3 Relationships between lacustrine sedimentary ancient plant DNA and vegetation around the lake and catchment areas. A, The composition of sedimentary plant DNA of high-altitude cryosphere lakes will be affected by DNA-free weathering substances (dilution effect). B, Egetation around the lakes has a high contribution to the lacustrine sedimentary plant DNA in the isolated lakes. C, Lake sediments may capture more plant DNA composition of different vegetation types in the larger catchment with developed hydrological conditions.
| 序号 Serial number | 研究地点 Study site | 材料来源 Source materials | 研究方法 Methodology | 物种检测率 Species detection rate | DNA结果分辨率 DNA resolution | 参考文献 Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 花粉 Pollen | 大化石 Macrofossil | DNA | 目 Order | 科 Family | 属 Genus | 种 Species | |||||
| 1 | 挪威 Norway | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | - | 20 | 16 | Parducci et al., | ||||
| 2 | 挪威瑞典边界 Norwegian-Swedish border | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 46 | - | 14 | Parducci et al., | ||||
| 3 | 格陵兰 Greenland | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 46 | 37 | 13 | Pedersen et al., | ||||
| 4 | 非洲 Africa | 泥炭、湖泊沉积物 Peat, Lacustrine sediments | 宏条形码 Metabarcoding | 26 | - | 34 | Boessenkool et al., | ||||
| 5 | 挪威 Norway | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 90 | - | 14 | Paus et al., | ||||
| 6 | 芬兰、俄罗斯西北部 Finland, Northwest Russian | 泥炭 Peat | 宏条形码 Metabarcoding | 54 | 36 | 10 | Parducci et al., | ||||
| 7 | 挪威 Norway | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | - | 30 | 34 | 13 | 32 | 19 | Alsos et al., | |
| 8 | 格陵兰 Greenland | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | - | - | 17 | 1 | 2 | 7 | 7 | Epp et al., |
| 9 | 阿尔卑斯山 Alps | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | - | - | 43 | 30 | 38 | 9 | Pansu et al., | |
| 10 | 白令海峡 Bering Strait | 湖泊沉积物 Lacustrine sediments | 宏基因组学 Metagenomics | 32 | 18 | 40 | Pedersen et al., | ||||
| 11 | 白令海峡 Bering Strait | 湖泊沉积物 Lacustrine sediments | 宏基因组学 Metagenomics | 64 | 22 | 55 | Pedersen et al., | ||||
| 12 | 非洲 Africa | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | >100 | - | 42 | 22 | 21 | Bremond et al., | ||
| 13 | 西伯利亚 Siberia | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 43 | - | 114 | 12 | 50 | 52 | Niemeyer et al., | |
| 14 | 苏格兰 Scotland | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | - | - | 73 | Sjögren et al., | ||||
| 15 | 西伯利亚 Siberia | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 84 | - | 113 | 1 | 22 | 49 | 40 | Zimmermann et al., |
| 16 | 挪威 Norway | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | - | - | 81 | Alsos et al., | ||||
| 17 | 挪威 Norway | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 39 | - | 118 | 14 | 55 | 49 | Clarke et al., | |
| 18 | 瑞典 Sweden | 泥炭 Peat | 宏基因组学 Metagenomics | 44 | 22 | 51 | 13 | 11 | 16 | 11 | Parducci et al., |
| 19 | 极地乌拉尔 Polar Urals | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 115 | - | 134 | 20 | 60 | 54 | Clarke et al., | |
| 20 | 非洲乌干达 Uganda, Africa | 泥炭 Peat | 宏基因组学 Metagenomics | - | - | 21 | Dommain et al., | ||||
| 21 | 西伯利亚 Siberia | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 52 | - | 120 | 42 | Liu et al., | |||
| 22 | 西伯利亚 Siberia | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 59 | - | 117 | 45 | Liu et al., | |||
| 23 | 西伯利亚 Siberia | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 55 | - | 90 | 34 | Liu et al., | |||
Table 1 Comparison of lacustrine (peat) plant sedimentary ancient DNA with pollen and macrofossil, and the species resolution revealed by plant DNA*
| 序号 Serial number | 研究地点 Study site | 材料来源 Source materials | 研究方法 Methodology | 物种检测率 Species detection rate | DNA结果分辨率 DNA resolution | 参考文献 Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 花粉 Pollen | 大化石 Macrofossil | DNA | 目 Order | 科 Family | 属 Genus | 种 Species | |||||
| 1 | 挪威 Norway | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | - | 20 | 16 | Parducci et al., | ||||
| 2 | 挪威瑞典边界 Norwegian-Swedish border | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 46 | - | 14 | Parducci et al., | ||||
| 3 | 格陵兰 Greenland | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 46 | 37 | 13 | Pedersen et al., | ||||
| 4 | 非洲 Africa | 泥炭、湖泊沉积物 Peat, Lacustrine sediments | 宏条形码 Metabarcoding | 26 | - | 34 | Boessenkool et al., | ||||
| 5 | 挪威 Norway | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 90 | - | 14 | Paus et al., | ||||
| 6 | 芬兰、俄罗斯西北部 Finland, Northwest Russian | 泥炭 Peat | 宏条形码 Metabarcoding | 54 | 36 | 10 | Parducci et al., | ||||
| 7 | 挪威 Norway | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | - | 30 | 34 | 13 | 32 | 19 | Alsos et al., | |
| 8 | 格陵兰 Greenland | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | - | - | 17 | 1 | 2 | 7 | 7 | Epp et al., |
| 9 | 阿尔卑斯山 Alps | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | - | - | 43 | 30 | 38 | 9 | Pansu et al., | |
| 10 | 白令海峡 Bering Strait | 湖泊沉积物 Lacustrine sediments | 宏基因组学 Metagenomics | 32 | 18 | 40 | Pedersen et al., | ||||
| 11 | 白令海峡 Bering Strait | 湖泊沉积物 Lacustrine sediments | 宏基因组学 Metagenomics | 64 | 22 | 55 | Pedersen et al., | ||||
| 12 | 非洲 Africa | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | >100 | - | 42 | 22 | 21 | Bremond et al., | ||
| 13 | 西伯利亚 Siberia | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 43 | - | 114 | 12 | 50 | 52 | Niemeyer et al., | |
| 14 | 苏格兰 Scotland | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | - | - | 73 | Sjögren et al., | ||||
| 15 | 西伯利亚 Siberia | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 84 | - | 113 | 1 | 22 | 49 | 40 | Zimmermann et al., |
| 16 | 挪威 Norway | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | - | - | 81 | Alsos et al., | ||||
| 17 | 挪威 Norway | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 39 | - | 118 | 14 | 55 | 49 | Clarke et al., | |
| 18 | 瑞典 Sweden | 泥炭 Peat | 宏基因组学 Metagenomics | 44 | 22 | 51 | 13 | 11 | 16 | 11 | Parducci et al., |
| 19 | 极地乌拉尔 Polar Urals | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 115 | - | 134 | 20 | 60 | 54 | Clarke et al., | |
| 20 | 非洲乌干达 Uganda, Africa | 泥炭 Peat | 宏基因组学 Metagenomics | - | - | 21 | Dommain et al., | ||||
| 21 | 西伯利亚 Siberia | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 52 | - | 120 | 42 | Liu et al., | |||
| 22 | 西伯利亚 Siberia | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 59 | - | 117 | 45 | Liu et al., | |||
| 23 | 西伯利亚 Siberia | 湖泊沉积物 Lacustrine sediments | 宏条形码 Metabarcoding | 55 | - | 90 | 34 | Liu et al., | |||
| 对比方面 Contrast aspect | 花粉 Pollen | 植物古DNA Plant ancient DNA | |
|---|---|---|---|
| 现代过程 Modern process | 产率 Productivity | 已有相对花粉产率研究, 花粉R值 Relative pollen productivity and R-value | - |
| 来源范围 Source area | 区域尺度 Regional-scale | 流域尺度、湖泊周边 Catchment scale and around the lake | |
| 传播途径 Dispersal route | 风力传播为主, 兼有流水搬运、昆虫携带和坡面侵蚀等 Anemophilous, water transportation, entomophilous, and slope erosion pathway | 风力传播较弱, 流水搬运更强, 兼有昆虫携带和坡面侵蚀等 Weaker anemophilous, stronger water transportation, entomophilous and slope erosion pathway | |
| 损伤机制 Damage mechanism | 机械破碎和化学溶蚀 Mechanical disruption and chemical corrosion | 普遍发生的DNA降解, 如脱嘌呤作用、水解作用和氧化作用以及微生物分解 Common DNA degradation, such as depurination, hydrolysis, oxidation and microbial decomposition | |
| 致损原因 Cause of damage | 仅在部分特殊环境下才导致花粉严重损伤, 如高pH或强水动力条件 Only in some special conditions, such as high pH or strong hydrodynamic conditions | 紫外线、pH、微生物、温度、水分等 UV, pH, microorganisms, temperature, moisture, etc | |
| 埋藏后过程 Post burial process | 表层扰动 Surface disturbance | 表层扰动、DNA降解 Surface disturbance, DNA degradation | |
| 埋藏时间 Burial time | 第四纪甚至更久 ≥Quaternary | 小于10万年 <100 000 years | |
| 与现代植被的关系 Relationship with modern vegetation | 物种分辨率 Species resolution | 科、属, 部分到种 Family, genus, partly to species | 科、属、种、亚种 Family, genus, species, subspecies |
| 植物代表性 Plant representativeness | 花粉R值等 Pollen R-value, etc | 仅部分结果支持DNA丰度与现代植被丰度正相关 Only a few results support the positive correlation | |
| 重建古植被 Palaeovegetation reconstruction | BIOME和REVEALS模型等 BIOME and REVEALS model etc | - | |
| 植物多样性研究 Relevant studies | 花粉多样性和植被多样性 Pollen diversity and vegetation diversity | 植物多样性, 能够揭示更多的稀有种或本地种 Plant diversity, reveal more rare or native species | |
| 其他 Else | 研究过程 Research process | 依赖分类专家, 鉴定结果耗时较长 Needs taxonomists and takes a long time | 依赖实验操作流程的质量控制, 包括污染控制、数据真实性验证等; 依赖现代植物分子数据库 Not only relies on the operation quality control, including pollution control and data authenticity verification but also needs the modern plant molecular database |
| 应用 Application | 局地或区域植物群落变化, 植被类型变化; 标准化, 可进行远距离的研究结果对比, 已用于探讨大尺度的植被演变、古气候变化等科学问题 Changes of local or regional plant communities and vegetation types. Standardization can be used to compare different research results and has been used to discuss large-scale vegetation evolution, palaeoclimate change, and other scientific problems | 植物物种发育关系, 局地植物群落变化, 植被类型和植物多样性变化; 方法个性化导致不同研究结果无法直接对比, 或者存在较大的不确定性 Phylogenetic relationships of plant species, changes of local plant communities, vegetation types, and plant diversity. The individuation of methods leads to the inability of direct comparison between different research results, or the existence of great uncertainty |
Table 2 Comparison of pollen and ancient plant DNA
| 对比方面 Contrast aspect | 花粉 Pollen | 植物古DNA Plant ancient DNA | |
|---|---|---|---|
| 现代过程 Modern process | 产率 Productivity | 已有相对花粉产率研究, 花粉R值 Relative pollen productivity and R-value | - |
| 来源范围 Source area | 区域尺度 Regional-scale | 流域尺度、湖泊周边 Catchment scale and around the lake | |
| 传播途径 Dispersal route | 风力传播为主, 兼有流水搬运、昆虫携带和坡面侵蚀等 Anemophilous, water transportation, entomophilous, and slope erosion pathway | 风力传播较弱, 流水搬运更强, 兼有昆虫携带和坡面侵蚀等 Weaker anemophilous, stronger water transportation, entomophilous and slope erosion pathway | |
| 损伤机制 Damage mechanism | 机械破碎和化学溶蚀 Mechanical disruption and chemical corrosion | 普遍发生的DNA降解, 如脱嘌呤作用、水解作用和氧化作用以及微生物分解 Common DNA degradation, such as depurination, hydrolysis, oxidation and microbial decomposition | |
| 致损原因 Cause of damage | 仅在部分特殊环境下才导致花粉严重损伤, 如高pH或强水动力条件 Only in some special conditions, such as high pH or strong hydrodynamic conditions | 紫外线、pH、微生物、温度、水分等 UV, pH, microorganisms, temperature, moisture, etc | |
| 埋藏后过程 Post burial process | 表层扰动 Surface disturbance | 表层扰动、DNA降解 Surface disturbance, DNA degradation | |
| 埋藏时间 Burial time | 第四纪甚至更久 ≥Quaternary | 小于10万年 <100 000 years | |
| 与现代植被的关系 Relationship with modern vegetation | 物种分辨率 Species resolution | 科、属, 部分到种 Family, genus, partly to species | 科、属、种、亚种 Family, genus, species, subspecies |
| 植物代表性 Plant representativeness | 花粉R值等 Pollen R-value, etc | 仅部分结果支持DNA丰度与现代植被丰度正相关 Only a few results support the positive correlation | |
| 重建古植被 Palaeovegetation reconstruction | BIOME和REVEALS模型等 BIOME and REVEALS model etc | - | |
| 植物多样性研究 Relevant studies | 花粉多样性和植被多样性 Pollen diversity and vegetation diversity | 植物多样性, 能够揭示更多的稀有种或本地种 Plant diversity, reveal more rare or native species | |
| 其他 Else | 研究过程 Research process | 依赖分类专家, 鉴定结果耗时较长 Needs taxonomists and takes a long time | 依赖实验操作流程的质量控制, 包括污染控制、数据真实性验证等; 依赖现代植物分子数据库 Not only relies on the operation quality control, including pollution control and data authenticity verification but also needs the modern plant molecular database |
| 应用 Application | 局地或区域植物群落变化, 植被类型变化; 标准化, 可进行远距离的研究结果对比, 已用于探讨大尺度的植被演变、古气候变化等科学问题 Changes of local or regional plant communities and vegetation types. Standardization can be used to compare different research results and has been used to discuss large-scale vegetation evolution, palaeoclimate change, and other scientific problems | 植物物种发育关系, 局地植物群落变化, 植被类型和植物多样性变化; 方法个性化导致不同研究结果无法直接对比, 或者存在较大的不确定性 Phylogenetic relationships of plant species, changes of local plant communities, vegetation types, and plant diversity. The individuation of methods leads to the inability of direct comparison between different research results, or the existence of great uncertainty |
| [1] |
Allen JRM, Huntley B (1999). Estimating past floristic diversity in montane regions from macrofossil assemblages. Journal of Biogeography, 26, 55-73.
DOI URL |
| [2] |
Alsos IG, Lammers Y, Yoccoz NG, Jørgensen T, Sjögren P, Gielly L, Edwards ME (2018). Plant DNA metabarcoding of lake sediments: How does it represent the contemporary vegetation. PLOS ONE, 13, e0195403. DOI: 10.1371/journal.pone.0195403.
DOI URL |
| [3] |
Alsos IG, Sjögren P, Edwards ME, Landvik JY, Gielly L, Forwick M, Coissac E, Brown AG, Jakobsen LV, Føreid MK, Pedersen MW (2016). Sedimentary ancient DNA from Lake Skartjørna, Svalbard: assessing the resilience of arctic flora to Holocene climate change. Holocene, 26, 627- 642.
DOI URL |
| [4] |
Anderson-Carpenter LL, McLachlan JS, Jackson ST, Kuch M, Lumibao CY, Poinar HN (2011). Ancient DNA from lake sediments: bridging the gap between paleoecology and genetics. BMC Evolutionary Biology, 11, 30. DOI: 10.1186/1471-2148-11-30
DOI PMID |
| [5] |
Bajard M, Poulenard J, Sabatier P, Etienne D, Ficetola F, Chen W, Gielly L, Taberlet P, Develle AL, Rey PJ, Moulin B, de Beaulieu JL, Arnaud F (2017). Long-term changes in alpine pedogenetic processes: effect of millennial agro- pastoralism activities (French-Italian Alps). Geoderma, 306, 217-236.
DOI URL |
| [6] |
Banchi E, Ametrano CG, Tordoni E, Stanković D, Ongaro S, Tretiach M, Pallavicini A, Muggia L, Group ARPAW (2020). Environmental DNA assessment of airborne plant and fungal seasonal diversity. Science of the Total Environment, 738, 140249. DOI: 10.1016/j.scitotenv.2020.140249.
DOI URL |
| [7] |
Barnes MA, Turner CR (2016). The ecology of environmental DNA and implications for conservation genetics. Conservation Genetics, 17, 1-17.
DOI URL |
| [8] |
Bennett KD, Parducci L (2006). DNA from pollen: principles and potential. Holocene, 16, 1031-1034.
DOI URL |
| [9] |
Birks HJB, Berglund BE (2018). One hundred years of Quaternary pollen analysis 1916-2016. Vegetation History and Archaeobotany, 27, 271-309.
DOI URL |
| [10] |
Birks HJB, Birks HH (2016). How have studies of ancient DNA from sediments contributed to the reconstruction of Quaternary floras? New Phytologist, 209, 499-506.
DOI PMID |
| [11] |
Blum SAE, Lorenz MG, Wackernagel W (1997). Mechanism of retarded DNA degradation and prokaryotic origin of DNases in nonsterile soils. Systematic and Applied Microbiology, 20, 513-521.
DOI URL |
| [12] |
Boessenkool S, McGlynn G, Epp LS, Taylor D, Pimentel M, Gizaw A, Nemomissa S, Brochmann C, Popp M (2014). Use of ancient sedimentary DNA as a novel conservation tool for high-altitude tropical biodiversity. Conservation Biology, 28, 446-455.
DOI PMID |
| [13] |
Bremond L, Favier C, Ficetola GF, Tossou MG, Akouégninou A, Gielly L, Giguet-Covex C, Oslisly R, Salzmann U (2017). Five thousand years of tropical lake sediment DNA records from Benin. Quaternary Science Reviews, 170, 203-211.
DOI URL |
| [14] |
Brown TA, Barnes IM (2015). The current and future applications of ancient DNA in Quaternary science. Journal of Quaternary Science, 30, 144-153.
DOI URL |
| [15] |
Clarke CL, Alsos IG, Edwards ME, Paus A, Gielly L, Haflidason H, Mangerud J, Regnéll C, Hughes PDM, Svendsen JI, Bjune AE (2020). A 24,000-year ancient DNA and pollen record from the Polar Urals reveals temporal dynamics of Arctic and boreal plant communities. Quaternary Science Reviews, 247, 106564. DOI: 10.1016/j.quascirev.2020.106564.
DOI URL |
| [16] |
Clarke CL, Edwards ME, Brown AG, Gielly L, Lammers Y, Heintzman PD, Ancin-Murguzur FJ, Brathen KA, Goslar T, Alsos IG (2019a). Holocene floristic diversity and richness in northeast Norway revealed by sedimentary ancient DNA (sedaDNA) and pollen. Boreas, 48, 299-316.
DOI URL |
| [17] |
Clarke CL, Edwards ME, Gielly L, Ehrich D, Hughes PDM, Morozova LM, Haflidason H, Mangerud J, Svendsen JI, Alsos IG (2019b). Persistence of arctic-alpine flora during 24,000 years of environmental change in the Polar Urals. Scientific Reports, 9, 19613. DOI: 10.1038/s41598-019-55989-9.
DOI |
| [18] |
Corinaldesi C, Beolchini F, Dell'anno A (2008). Damage and degradation rates of extracellular DNA in marine sediments: implications for the preservation of gene sequences. Molecular Ecology, 17, 3939-3951.
DOI PMID |
| [19] |
Crump SE, Miller GH, Power M, Sepúlveda J, Dildar N, Coghlan M, Bunce M (2019). Arctic shrub colonization lagged peak postglacial warmth: molecular evidence in lake sediment from Arctic Canada. Global Change Biology, 25, 4244-4256.
DOI URL |
| [20] |
Demanèche S, Jocteur-Monrozier L, Quiquampoix H, Simonet P (2001). Evaluation of biological and physical protection against nuclease degradation of clay-bound plasmid DNA. Applied and Environmental Microbiology, 67, 293-299.
PMID |
| [21] |
Dommain R, Andama M, McDonough MM, Prado NA, Goldhammer T, Potts R, Maldonado JE, Nkurunungi JB, Campana MG (2020). The challenges of reconstructing tropical biodiversity with sedimentary ancient DNA: a 2200-year- long metagenomic record from bwindi impenetrable forest, Uganda. Frontiers in Ecology and Evolution, 8, 218. DOI: 10.3389/fevo.2020.00218.
DOI URL |
| [22] |
Dougherty MM, Larson ER, Renshaw MA, Gantz CA, Egan SP, Erickson DM, Lodge DM (2016). Environmental DNA (eDNA) detects the invasive rusty crayfish Orconectes rusticus at low abundances. Journal of Applied Ecology, 53, 722-732.
PMID |
| [23] | Dulias K, Knockaert J, Giguet-Covex C, Walsh K (2020). The use of sedimentary ancient DNA from lakes in tracing human-environment interactions in the Western Alps [2021-10-28]. https://meetingorganizer.copernicus.org/EGU2020/EGU2020-21944.html. |
| [24] |
Dunker KJ, Sepulveda AJ, Massengill RL, Olsen JB, Russ OL, Wenburg JK, Antonovich A (2016). Potential of environmental DNA to evaluate northern pike (Esox lucius) eradication efforts: an experimental test and case study. PLOS ONE, 11, e0162277. DOI: 10.1371/journal.pone.0162277.
DOI URL |
| [25] |
Edwards ME (2020). The maturing relationship between Quaternary paleoecology and ancient sedimentary DNA. Quaternary Research, 96, 39-47.
DOI URL |
| [26] |
Edwards ME, Alsos IG, Yoccoz N, Coissac E, Goslar T, Gielly L, Haile J, Langdon CT, Tribsch A, Binney HA, von Stedingk H, Taberlet P (2018). Metabarcoding of modern soil DNA gives a highly local vegetation signal in Svalbard tundra. The Holocene, 28, 2006-2016.
DOI URL |
| [27] |
Eichmiller JJ, Best SE, Sorensen PW (2016). Effects of temperature and trophic state on degradation of environmental DNA in lake water. Environmental Science & Technology, 50, 1859-1867.
DOI URL |
| [28] | Ellis JR, Bentley KE, McCauley DE (2008). Detection of rare paternal chloroplast inheritance in controlled crosses of the endangered sunflower Helianthus verticillatus. Heredity, 100, 574-580. |
| [29] |
Epp LS, Gussarova G, Boessenkool S, Olsen J, Haile J, Schrøder-Nielsen A, Ludikova A, Hassel K, Stenøien HK, Funder S, Willerslev E, Kjær K, Brochmann C (2015). Lake sediment multi-taxon DNA from North Greenland records early post-glacial appearance of vascular plants and accurately tracks environmental changes. Quaternary Science Reviews, 117, 152-163.
DOI URL |
| [30] |
Evrard O, Laceby JP, Ficetola GF, Gielly L, Huon S, Lefèvre I, Onda Y, Poulenard J (2019). Environmental DNA provides information on sediment sources: a study in catchments affected by Fukushima radioactive fallout. Science of the Total Environment, 665, 873-881.
DOI URL |
| [31] |
Ficetola GF, Manenti R, Taberlet P (2019). Environmental DNA and metabarcoding for the study of amphibians and reptiles: species distribution, the microbiome, and much more. Amphibia-Reptilia, 40, 129-148.
DOI URL |
| [32] |
Ficetola GF, Poulenard J, Sabatier P, Messager E, Gielly L, Leloup A, Etienne D, Bakke J, Malet E, Fanget B, Støren E, Reyss JL, Taberlet P, Arnaud F (2018). DNA from lake sediments reveals long-term ecosystem changes after a biological invasion. Science Advances, 4, eaar4292. DOI: 10.1126/sciadv.aar4292.
DOI URL |
| [33] | Fu XH, Wang L, Le YQ, Hu JJ (2012). Persistence and renaturation efficiency of thermally treated waste recombinant DNA in defined aquatic microcosms. Journal of Environmental Science and Health. Part A, Toxic/Hazardous Substances & Environmental Engineering, 47, 1975-1983. |
| [34] |
Giguet-Covex C, Ficetola GF, Walsh K, Poulenard J, Bajard M, Fouinat L, Sabatier P, Gielly L, Messager E, Develle AL, David F, Taberlet P, Brisset E, Guiter F, Sinet R, Arnaud F (2019). New insights on lake sediment DNA from the catchment: importance of taphonomic and analytical issues on the record quality. Scientific Reports, 9, 14676. DOI: 10.1038/s41598-019-50339-1.
DOI PMID |
| [35] |
Giguet-Covex C, Pansu J, Arnaud F, Rey PJ, Griggo C, Gielly L, Domaizon I, Coissac E, David F, Choler P, Poulenard J, Taberlet P (2014). Long livestock farming history and human landscape shaping revealed by lake sediment DNA. Nature Communications, 5, 3211. DOI: 10.1038/ncomms4211.
DOI PMID |
| [36] |
Greaves MP, Wilson MJ (1969). The adsorption of nucleic acids by montmorillonite. Soil Biology & Biochemistry, 1, 317-323.
DOI URL |
| [37] | Gu ZQ, Wang YC, Liu YQ, Yang XY, Chen FH (2020). Ancient environmental DNA and environmental archaeology. Quaternary Sciences, 40, 295-296. |
| [顾政权, 王昱程, 刘勇勤, 杨晓燕, 陈发虎 (2020). 古环境DNA与环境考古. 第四纪研究, 40, 295-296.] | |
| [38] |
Guo Y, Yang GQ, Chen YM, Li DZ, Guo ZH (2018). A comparison of different methods for preserving plant molecular materials and the effect of degraded DNA on ddRAD sequencing. Plant Diversity, 40, 106-116.
DOI URL |
| [39] | Häder DP, Kumar HD, Smith RC, Worrest RC (2003). Aquatic ecosystems: effects of solar ultraviolet radiation and interactions with other climatic change factors. Photochemical & Photobiological Sciences, 2, 39-50. |
| [40] |
Hawkins J, de Vere N, Griffith A, Ford CR, Allainguillaume J, Hegarty MJ, Baillie L, Adams-Groom B (2015). Using DNA metabarcoding to identify the floral composition of honey: a new tool for investigating honey bee foraging preferences. PLOS ONE, 10, e0134735. DOI: 10.1371/journal.pone.0134735.
DOI URL |
| [41] |
Heinecke L, Epp LS, Reschke M, Stoof-Leichsenring KR, Mischke S, Plessen B, Herzschuh U (2017). Aquatic macrophyte dynamics in Lake Karakul (Eastern Pamir) over the last 29 calka revealed by sedimentary ancient DNA and geochemical analyses of macrofossil remains. Journal of Paleolimnology, 58, 403-417.
DOI URL |
| [42] |
Hofreiter M, Serre D, Poinar HN, Kuch M, Pääbo S (2001). Ancient DNA. Nature Reviews Genetics, 2, 353-359.
PMID |
| [43] | Jia WH (2020). Modern-process Studies of Plant DNA in Lake Sediments, Qinghai-Tibetan Plateau and Arid Northwestern China. Master degree dissertation, Capital Normal University, Beijing. |
| [贾伟瀚 (2020). 青藏高原和西北干旱区湖泊沉积物中植物DNA的现代过程研究. 硕士学位论文, 首都师范大学, 北京.] | |
| [44] |
Jia WH, Liu XQ, Stoof-Leichsenring KR, Liu SS, Li K, Herzschuh U (2022). Preservation of sedimentary plant DNA is related to lake water chemistry. Environmental DNA, 4, 425-439.
DOI URL |
| [45] |
Jørgensen T, Haile J, Möller P, Andreev A, Boessenkool S, Rasmussen M, Kienast F, Coissac E, Taberlet P, Brochmann C, Bigelow NH, Andersen K, Orlando L, Gilbert MTP, Willerslev E (2012a). A comparative study of ancient sedimentary DNA, pollen and macrofossils from permafrost sediments of northern Siberia reveals long-term vegetational stability. Molecular Ecology, 21, 1989-2003.
DOI URL |
| [46] |
Jørgensen T, Kjaer KH, Haile J, Rasmussen M, Boessenkool S, Andersen K, Coissac E, Taberlet P, Brochmann C, Orlando L, Gilbert MTP, Willerslev E (2012b). Islands in the ice: detecting past vegetation on Greenlandic nunataks using historical records and sedimentary ancient DNA meta- barcoding. Molecular Ecology, 21, 1980-1988.
DOI URL |
| [47] |
Kanbar HJ, Olajos F, Englund G, Holmboe M (2020). Geochemical identification of potential DNA-hotspots and DNA-infrared fingerprints in lake sediments. Applied Geochemistry, 122, 104728. DOI: 10.1016/j.apgeochem.2020.104728
DOI URL |
| [48] |
Khanna M, Stotzky G (1992). Transformation of Bacillus subtilis by DNA bound on montmorillonite and effect of DNase on the transforming ability of bound DNA. Applied and Environmental Microbiology, 58, 1930-1939.
DOI PMID |
| [49] |
Kraaijeveld K, de Weger LA, Ventayol García M, Buermans H, Frank J, Hiemstra PS, den Dunnen JT (2015). Efficient and sensitive identification and quantification of airborne pollen using next-generation DNA sequencing. Molecular Ecology Resources, 15, 8-16.
DOI PMID |
| [50] | Kruse S, Epp LS, Wieczorek M, Pestryakova LA, Stoof- Leichsenring KR, Herzschuh U (2018). High gene flow and complex treeline dynamics of Larix Mill. stands on the Taymyr Peninsula (north-central Siberia) revealed by nuclear microsatellites. Tree Genetics & Genomes, 14, 1-14. |
| [51] |
Kruszewska H, Misicka A, Chmielowiec U (2004). Biodegradation of DNA and nucleotides to nucleosides and free bases. Il Farmaco, 59, 13-20.
DOI URL |
| [52] |
Lammers Y, Heintzman PD, Alsos IG (2021). Environmental palaeogenomic reconstruction of an Ice Age algal population. Communications Biology, 4, 220. DOI: 10.1038/s42003-021-01710-4.
DOI |
| [53] | Lan TY, Lindqvist C (2018). Paleogenomics:genome-scale analysis of ancient DNA and population and evolutionary genomic inferences//Rajora OP. Population Genomics: Concepts, Approaches and Applications. Springer, Cham, Switzerland. 323-360. |
| [54] |
Lear G, Dickie I, Banks J, Boyer S, Buckley H, Buckley T, Cruickshank R, Dopheide A, Handley K, Hermans S, Kamke J, Lee C, MacDiarmid R, Morales S, Orlovich D, et al. (2018). Methods for the extraction, storage, amplification and sequencing of DNA from environmental samples. New Zealand Journal of Ecology, 42, 10. DOI: 10.20417/nzjecol.42.9.
DOI |
| [55] |
Leontidou K, Vernesi C, Groeve J, Cristofolini F, Vokou D, Cristofori A (2018). DNA metabarcoding of airborne pollen: new protocols for improved taxonomic identification of environmental samples. Aerobiologia, 34, 63-74.
DOI URL |
| [56] |
Levy-Booth DJ, Campbell RG, Gulden RH, Hart MM, Powell JR, Klironomos JN, Pauls KP, Swanton CJ, Trevors JT, Dunfield KE (2007). Cycling of extracellular DNA in the soil environment. Soil Biology & Biochemistry, 39, 2977-2991.
DOI URL |
| [57] |
Li DZ, Wang YH, Yi TS, Wang H, Gao LM, Yang JB (2012). The next-generation flora: iFlora. Plant Diversity and Resources, 34, 525-531.
DOI URL |
| [李德铢, 王雨华, 伊廷双, 王红, 高连明, 杨俊波 (2012). 新一代植物志: iFlora. 植物分类与资源学报, 34, 525-531.] | |
| [58] |
Li K, Stoof-Leichsenring KR, Liu SS, Jia WH, Liao MN, Liu XQ, Ni J, Herzschuh U (2021). Plant sedimentary DNA as a proxy for vegetation reconstruction in eastern and northern Asia. Ecological Indicators, 132, 108303. DOI: 10.1016/j.ecolind.2021.108303.
DOI URL |
| [59] |
Lindahl T (1993). Instability and decay of the primary structure of DNA. Nature, 362, 709-715.
DOI URL |
| [60] |
Liu S, Kruse S, Scherler D, Ree RH, Zimmermann HH, Stoof-Leichsenring KR, Epp LS, Mischke S, Herzschuh U (2021a). Sedimentary ancient DNA reveals a threat of warming-induced alpine habitat loss to Tibetan Plateau plant diversity. Nature Communications, 12, 2995. DOI: 10.1038/s41467-021-22986-4.
DOI URL |
| [61] |
Liu SS, Li K, Jia WH, Stoof-Leichsenring KR, Liu XQ, Cao XY, Herzschuh U (2021b). Vegetation reconstruction from Siberia and the Tibetan Plateau using modern analogue technique-Comparing sedimentary (ancient) DNA and pollen data. Frontiers in Ecology and Evolution, 9, 668611. DOI: 10.3389/fevo.2021.668611.
DOI URL |
| [62] |
Liu SS, Stoof-Leichsenring KR, Kruse S, Pestryakova LA, Herzschuh U (2020). Holocene vegetation and plant diversity changes in the north-eastern Siberian treeline region from pollen and sedimentary ancient DNA. Frontiers in Ecology and Evolution, 8, 560243. DOI: 10.3389/fevo.2020.560243.
DOI URL |
| [63] | Ma RF, Zhang W, Liu L, Yang CY (2020). Application of sedimentary plant ancient DNA (aDNA) technology in paleovegetation reconstruction. Acta Sedimentologica Sinica, 38, 1179-1191. |
| [马瑞丰, 张威, 刘亮, 杨蝉玉 (2020). 沉积植物古DNA技术在古植被重建中的应用. 沉积学报, 38, 1179-1191.] | |
| [64] | MacHugh DE, Edwards CJ, Bailey JF, Bancroft DR, Bradley DG (2000). The extraction and analysis of ancient DNA from bone and teeth: a survey of current methodologies. Ancient Biomolecules, 3, 81-102. |
| [65] |
Magyari EK, Major A, Bálint M, Nédli J, Braun M, Rácz I, Parducci L (2011). Population dynamics and genetic changes of Picea abies in the South Carpathians revealed by pollen and ancient DNA analyses. BMC Evolutionary Biology, 11, 66. DOI: 10.1186/1471-2148-11-66.
DOI PMID |
| [66] |
Maruyama A, Nakamura K, Yamanaka H, Kondoh M, Minamoto T (2014). The release rate of environmental DNA from juvenile and adult fish. PLOS ONE, 9, e114639. DOI: 10.1371/journal.pone.0114639.
DOI URL |
| [67] |
Matesanz S, Pescador DS, Pías B, Sánchez AM, Chacón- Labella J, Illuminati A, de la Cruz M, López-Angulo J, Marí-Mena N, Vizcaíno A, Escudero A (2019). Estimating belowground plant abundance with DNA metabarcoding. Molecular Ecology Resources, 19, 1265-1277.
DOI PMID |
| [68] |
Matisoo-Smith E, Roberts K, Welikala N, Tannock G, Chester P, Feek D, Flenley J (2008). Recovery of DNA and pollen from New Zealand Lake sediments. Quaternary International, 184, 139-149.
DOI URL |
| [69] |
Murchie TJ, Kuch M, Duggan AT, Ledger ML, Roche K, Klunk J, Karpinski E, Hackenberger D, Sadoway T, MacPhee R, Froese D, Poinar H (2021). Optimizing extraction and targeted capture of ancient environmental DNA for reconstructing past environments using the PalaeoChip Arctic-1.0 bait-set. Quaternary Research, 99, 305-328.
DOI URL |
| [70] |
Murray DC, Pearson SG, Fullagar R, Chase BM, Houston J, Atchison J, White NE, Bellgard MI, Clarke E, Macphail M, Gilbert MTP, Haile J, Bunce M (2012). High- throughput sequencing of ancient plant and mammal DNA preserved in herbivore middens. Quaternary Science Reviews, 58, 135-145.
DOI URL |
| [71] |
Nielsen KM, Johnsen PJ, Bensasson D, Daffonchio D (2007). Release and persistence of extracellular DNA in the environment. Environmental Biosafety Research, 6, 37-53.
PMID |
| [72] | Niemeyer B, Epp LS, Stoof-Leichsenring KR, Pestryakova LA, Herzschuh U (2017). A comparison of sedimentary DNA and pollen from lake sediments in recording vegetation composition at the Siberian treeline. Molecular Ecology Resources, 17, e46-e62. |
| [73] | Núñez A, Amo de Paz G, Rastrojo A, García AM, Alcamí A, Gutiérrez-Bustillo AM, Moreno DA (2016). Monitoring of airborne biological particles in outdoor atmosphere. Part 2: Metagenomics applied to urban environments. International Microbiology, 19, 69-80. |
| [74] |
Ogram A, Sayler GS, Gustin D, Lewis RJ (1988). DNA adsorption to soils and sediments. Environmental Science & Technology, 22, 982-984.
DOI URL |
| [75] |
Pansu J, Giguet-Covex C, Ficetola GF, Gielly L, Boyer F, Zinger L, Arnaud F, Poulenard J, Taberlet P, Choler P (2015). Reconstructing long-term human impacts on plant communities: an ecological approach based on lake sediment DNA. Molecular Ecology, 24, 1485-1498.
DOI URL |
| [76] |
Parducci L, Alsos IG, Unneberg P, Pedersen MW, Han L, Lammers Y, Salonen JS, Väliranta MM, Slotte T, Wohlfarth B (2019). Shotgun environmental DNA, pollen, and macrofossil analysis of lateglacial lake sediments from southern Sweden. Frontiers in Ecology and Evolution, 7, 189. DOI: 10.3389/fevo.2019.00189.
DOI URL |
| [77] |
Parducci L, Bennett KD (2017). The real significance of ancient DNA. American Journal of Botany, 104, 800-802.
DOI PMID |
| [78] |
Parducci L, Bennett KD, Ficetola GF, Alsos IG, Suyama Y, Wood JR, Pedersen MW (2017). Ancient plant DNA in lake sediments. New Phytologist, 214, 924-942.
DOI PMID |
| [79] |
Parducci L, Jørgensen T, Tollefsrud MM, Elverland E, Alm T, Fontana SL, Bennett KD, Haile J, Matetovici I, Suyama Y, Edwards ME, Andersen K, Rasmussen M, Boessenkool S, Coissac E, et al. (2012). Glacial survival of boreal trees in northern Scandinavia. Science, 335, 1083-1086.
DOI PMID |
| [80] |
Parducci L, Matetovici I, Fontana SL, Bennett KD, Suyama Y, Haile J, Kjaer KH, Larsen NK, Drouzas AD, Willerslev E (2013). Molecular- and pollen-based vegetation analysis in lake sediments from central Scandinavia. Molecular Ecology, 22, 3511-3524.
DOI PMID |
| [81] | Parducci L, Nota K, Wood J (2018). Reconstructing past vegetation communities using ancient DNA from lake sediments// Lindqvist C, Rajora OP. Paleogenomics: Genome- Scale Analysis of Ancient DNA. Springer, Cham, Switzerland. 163-187. |
| [82] |
Parducci L, Petit RJ (2004). Ancient DNA-Unlocking plants' fossil secrets. New Phytologist, 161, 335-339.
DOI PMID |
| [83] |
Parducci L, Suyama Y, Lascoux M, Bennett KD (2005). Ancient DNA from pollen: a genetic record of population history in Scots pine. Molecular Ecology, 14, 2873-2882.
PMID |
| [84] |
Parducci L, Väliranta M, Salonen JS, Ronkainen T, Matetovici I, Fontana SL, Eskola T, Sarala P, Suyama Y (2015). Proxy comparison in ancient peat sediments: pollen, macrofossil and plant DNA. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 370, 20130382. DOI: 10.1098/rstb.2013.0382.
DOI |
| [85] |
Paus A, Boessenkool S, Brochmann C, Epp LS, Fabel D, Haflidason H, Linge H (2015). Lake Store Finnsjøen-A key for understanding Lateglacial/early Holocene vegetation and ice sheet dynamics in the central Scandes Mountains. Quaternary Science Reviews, 121, 36-51.
DOI URL |
| [86] |
Pedersen MW, Ginolhac A, Orlando L, Olsen J, Andersen K, Holm J, Funder S, Willerslev E, Kjær KH (2013). A comparative study of ancient environmental DNA to pollen and macrofossils from lake sediments reveals taxonomic overlap and additional plant taxa. Quaternary Science Reviews, 75, 161-168.
DOI URL |
| [87] |
Pedersen MW, Overballe-Petersen S, Ermini L, Sarkissian CD, Haile J, Hellstrom M, Spens J, Thomsen PF, Bohmann K, Cappellini E, Schnell IB, Wales NA, Carøe C, Campos PF, Schmidt AMZ, et al. (2015). Ancient and modern environmental DNA. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 370, 20130383. DOI: 10.1098/rstb.2013.0383.
DOI |
| [88] |
Pedersen MW, Ruter A, Schweger C, Friebe H, Staff RA, Kjeldsen KK, Mendoza MLZ, Beaudoin AB, Zutter C, Larsen NK, Potter BA, Nielsen R, Rainville RA, Orlando L, Meltzer DJ, et al. (2016). Postglacial viability and colonization in North America's ice-free corridor. Nature, 537, 45-49.
DOI URL |
| [89] |
Petersen G, Johansen B, Seberg O (1996). PCR and sequencing from a single pollen grain. Plant Molecular Biology, 31, 189-191.
PMID |
| [90] |
Pietramellara G, Ascher J, Borgogni F, Ceccherini MT, Guerri G, Nannipieri P (2009). Extracellular DNA in soil and sediment: fate and ecological relevance. Biology and Fertility of Soils, 45, 219-235.
DOI URL |
| [91] |
Pilliod DS, Goldberg CS, Arkle RS, Waits LP (2014). Factors influencing detection of eDNA from a stream-dwelling amphibian. Molecular Ecology Resources, 14, 109-116.
DOI PMID |
| [92] |
Poté J, Ackermann R, Wildi W (2009). Plant leaf mass loss and DNA release in freshwater sediments. Ecotoxicology and Environmental Safety, 72, 1378-1383.
DOI URL |
| [93] |
Potter C, de Vere N, Jones LE, Ford CR, Hegarty MJ, Hodder KH, Diaz A, Franklin EL (2019). Pollen metabarcoding reveals broad and species-specific resource use by urban bees. PeerJ, 7, e5999. DOI: 10.7717/peerj.5999.
DOI URL |
| [94] |
Prentice C, Guiot J, Huntley B, Jolly D, Cheddadi R (1996). Reconstructing biomes from palaeoecological data: a general method and its application to European pollen data at 0 and 6 ka. Climate Dynamics, 12, 185-194.
DOI URL |
| [95] |
Ravanat JL, Douki T, Cadet J (2001). Direct and indirect effects of UV radiation on DNA and its components. Journal of Photochemistry and Photobiology B: Biology, 63, 88-102.
DOI URL |
| [96] |
Rawlence NJ, Lowe DJ, Wood JR, Young JM, Churchman GJ, Huang YT, Cooper A (2014). Using palaeoenvironmental DNA to reconstruct past environments: progress and prospects. Journal of Quaternary Science, 29, 610-626.
DOI URL |
| [97] |
Rijal DP, Heintzman PD, Lammers Y, Yoccoz NG, Lorberau KE, Pitelkova I, Goslar T, Murguzur FJA, Salonen JS, Helmens KF, Bakke J, Edwards ME, Alm T, Bråthen KA, Brown AG, Alsos IG (2021). Sedimentary ancient DNA shows terrestrial plant richness continuously increased over the Holocene in northern Fennoscandia. Science Advances, 7, eabf9557. DOI: 10.1126/sciadv.abf9557.
DOI URL |
| [98] |
Romanowski G, Lorenz MG, Wackernagel W (1991). Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Applied and Environmental Microbiology, 57, 1057-1061.
DOI PMID |
| [99] |
Schlumbaum A, Tensen M, Jaenicke-Després V (2008). Ancient plant DNA in archaeobotany. Vegetation History and Archaeobotany, 17, 233-244.
DOI URL |
| [100] |
Schmid S, Genevest R, Gobet E, Suchan T, Sperisen C, Tinner W, Alvarez N (2017). HyRAD-X, a versatile method combining exome capture and RAD sequencing to extract genomic information from ancient DNA. Methods in Ecology and Evolution, 8, 1374-1388.
DOI URL |
| [101] |
Schulte L, Bernhardt N, Stoof-Leichsenring K, Zimmermann HH, Pestryakova LA, Epp LS, Herzschuh U (2021). Hybridization capture of larch (Larix Mill.) chloroplast genomes from sedimentary ancient DNA reveals past changes of Siberian forest. Molecular Ecology Resources, 21, 801-815.
DOI PMID |
| [102] |
Sheng X, Qin C, Yang B, Hu XJ, Liu C, Waigi MG, Li XL, Ling WT (2019). Metal cation saturation on montmorillonites facilitates the adsorption of DNA via cation bridging. Chemosphere, 235, 670-678.
DOI PMID |
| [103] |
Sirois SH, Buckley DH (2019). Factors governing extracellular DNA degradation dynamics in soil. Environmental Microbiology Reports, 11, 173-184.
DOI PMID |
| [104] |
Sjögren P, Edwards ME, Gielly L, Langdon CT, Croudace IW, Merkel MKF, Fonville T, Alsos IG (2017). Lake sedimentary DNA accurately records 20th century introductions of exotic conifers in Scotland. New Phytologist, 213, 929- 941.
DOI PMID |
| [105] |
Smith O, Momber G, Bates R, Garwood P, Fitch S, Pallen M, Gaffney V, Allaby RG (2015). Sedimentary DNA from a submerged site reveals wheat in the British Isles 8000 years ago. Science, 347, 998-1001.
DOI URL |
| [106] |
Sommaruga R (2001). The role of solar UV radiation in the ecology of alpine lakes. Journal of Photochemistry and Photobiology B: Biology, 62, 35-42.
DOI URL |
| [107] |
Sønstebø JH, Gielly L, Brysting AK, Elven R, Edwards M, Haile J, Willerslev E, Coissac E, Rioux D, Sannier J, Taberlet P, Brochmann C (2010). Using next-generation sequencing for molecular reconstruction of past Arctic vegetation and climate. Molecular Ecology Resources, 10, 1009-1018.
DOI PMID |
| [108] |
Stevens CJ, Fuller DQ (2012). Did Neolithic farming fail? The case for a Bronze Age agricultural revolution in the British Isles. Antiquity, 86, 707-722.
DOI URL |
| [109] |
Strickler KM, Fremier AK, Goldberg CS (2015). Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biological Conservation, 183, 85-92.
DOI URL |
| [110] |
Sugita S (2007a). Theory of quantitative reconstruction of vegetation I: Pollen from large sites REVEALS regional vegetation composition. The Holocene, 17, 229-241.
DOI URL |
| [111] |
Sugita S (2007b). Theory of quantitative reconstruction of vegetation II: All you need is LOVE. The Holocene, 17, 243-257.
DOI URL |
| [112] | Suyama Y, Kawamuro K, Kinoshita I, Yoshimura K, Tsumura Y, Takahara H (1996). DNA sequence from a fossil pollen of Abies spp. from Pleistocene peat. Genes & Genetic Systems, 71, 145-149. |
| [113] | Taberlet P, Bonin A, Zinger L, Coissac E (2018). Environmental DNA: for Biodiversity Research and Monitoring. Oxford Press, Oxford. |
| [114] |
Taberlet P, Coissac E, Hajibabaei M, Rieseberg LH (2012). Environmental DNA. Molecular Ecology, 21, 1789-1793.
DOI PMID |
| [115] |
Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, Vermat T, Corthier G, Brochmann C, Willerslev E (2006). Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Research, 35, e14. DOI: 10.1093/nar/gkl938.
DOI URL |
| [116] |
Varotto C, Pindo M, Bertoni E, Casarotto C, Camin F, Girardi M, Maggi V, Cristofori A (2021). A pilot study of eDNA metabarcoding to estimate plant biodiversity by an alpine glacier core (Adamello glacier, North Italy). Scientific Reports, 11, 1208. DOI: 10.1038/s41598-020-79738-5.
DOI URL |
| [117] |
Vasselon V, Bouchez A, Rimet F, Jacquet S, Trobajo R, Corniquel M, Tapolczai K, Domaizon I (2018). Avoiding quantification bias in metabarcoding: application of a cell biovolume correction factor in diatom molecular biomonitoring. Methods in Ecology and Evolution, 9, 1060-1069.
DOI URL |
| [118] |
Voldstad LH, Alsos IG, Farnsworth WR, Heintzman PD, Håkansson L, Kjellman SE, Rouillard A, Schomacker A, Eidesen PB (2020). A complete Holocene lake sediment ancient DNA record reveals long-standing high Arctic plant diversity hotspot in northern Svalbard. Quaternary Science Reviews, 234, 106207. DOI: 10.1016/j.quascirev.2020.106207.
DOI URL |
| [119] |
Wei N, Nakajima F, Tobino T (2018). A microcosm study of surface sediment environmental DNA: decay observation, abundance estimation, and fragment length comparison. Environmental Science & Technology, 52, 12428-12435.
DOI URL |
| [120] |
Willerslev E, Cappellini E, Boomsma W, Nielsen R, Hebsgaard MB, Brand TB, Hofreiter M, Bunce M, Poinar HN, Dahl-Jensen D, Johnsen S, Steffensen JP, Bennike O, Schwenninger JL, Nathan R, et al. (2007). Ancient biomolecules from deep ice cores reveal a forested southern Greenland. Science, 317, 111-114.
PMID |
| [121] | Willerslev E, Cooper A (2005). Ancient DNA. Proceedings. Biological Sciences, 272, 3-16. |
| [122] |
Willerslev E, Davison J, Moora M, Zobel M, Coissac E, Edwards ME, Lorenzen ED, Vestergård M, Gussarova G, Haile J, Craine J, Gielly L, Boessenkool S, Epp LS, Pearman PB, et al. (2014). Fifty thousand years of Arctic vegetation and megafaunal diet. Nature, 506, 47-51.
DOI URL |
| [123] |
Willerslev E, Hansen AJ, Binladen J, Brand TB, Gilbert MTP, Shapiro B, Bunce M, Wiuf C, Gilichinsky DA, Cooper A (2003). Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science, 300, 791- 795.
PMID |
| [124] |
Yoccoz NG, Bråthen KA, Gielly L, Haile J, Edwards ME, Goslar T, von Stedingk H, Brysting AK, Coissac E, Pompanon F, Sønstebø JH, Miquel C, Valentini A, de Bello F, Chave J, et al. (2012). DNA from soil mirrors plant taxonomic and growth form diversity. Molecular Ecology, 21, 3647-3655.
DOI PMID |
| [125] |
Zeng CX, Wang YN, Wang YH, Wang H (2014). Plant DNA barcoding and framework for biodiversity data sharing platform. Biodiversity Science, 22, 285-292.
DOI URL |
|
[曾春霞, 王亚楠, 王雨华, 王红 (2014). 植物DNA条形码与生物多样性数据共享平台构建. 生物多样性, 22, 285-292.]
DOI |
|
| [126] | Zhang DJ, Dong GH, Wang H, Ren XY, Ha P, Qiang MR, Chen FH (2016). History and possible mechanisms of prehistoric human migration to the Tibetan Plateau. Science China Earth Sciences, 46, 1007-1023. |
| [张东菊, 董广辉, 王辉, 任晓燕, 哈比布, 强明瑞, 陈发虎 (2016). 史前人类向青藏高原扩散的历史过程和可能驱动机制. 中国科学: 地球科学, 46, 1007-1023.] | |
| [127] |
Zhang Q, Liu Y, Sodmergen S (2003). Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant and Cell Physiology, 44, 941-951.
PMID |
| [128] |
Zhu B (2006). Degradation of plasmid and plant DNA in water microcosms monitored by natural transformation and real-time polymerase chain reaction (PCR). Water Research, 40, 3231-3238.
DOI URL |
| [129] |
Zimmermann HH, Raschke E, Epp LS, Stoof-Leichsenring KR, Schwamborn G, Schirrmeister L, Overduin PP, Herzschuh U (2017). Sedimentary ancient DNA and pollen reveal the composition of plant organic matter in Late Quaternary permafrost sediments of the Buor Khaya Peninsula (north-eastern Siberia). Biogeosciences, 14, 575-596.
DOI URL |
| [130] |
Zobel M, Davison J, Edwards ME, Brochmann C, Coissac E, Taberlet P, Willerslev E, Moora M (2018). Ancient environmental DNA reveals shifts in dominant mutualisms during the late Quaternary. Nature Communications, 9, 139. DOI: 10.1038/s41467-017-02421-3.
DOI URL |
| [131] |
Zolitschka B, Francus P, Ojala AEK, Schimmelmann A (2015). Varves in lake sediments-A review. Quaternary Science Reviews, 117, 1-41.
DOI URL |
| [1] | ZHOU Bo-Rui, LIAO Meng-Na, LI Kai, XU De-Yu, CHEN Hai-Yan, NI Jian, CAO Xian-Yong, KONG Zhao-Chen, XU Qing-Hai, ZHANG Yun, Ulrike HERZSCHUH, CAI Yong-Li, CHEN Bi-Shan, CHEN Jing-An, CHEN Ling-Kang, CHENG Bo, GAO Yang, $\boxed{\hbox{HUANG Ci-Xuan}}$ , HUANG Xiao-Zhong, LI Sheng-Feng, LI Wen-Yi, LIU Kam-Biu, LIU Guang-Xiu, LIU Ping-Mei, LIU Xing-Qi, MA Chun-Mei, SONG Chang-Qing, SUN Xiang-Jun, TANG Ling-Yu, WANG Man-Hua, WANG Yong-Bo, $\boxed{\hbox{XIA Yu-Mei}}$ , XU Jia-Sheng, YAN Shun, YANG Xiang-Dong, YAO Yi-Feng, YE Chuan-Yong, ZHANG Zhi-Yong, ZHAO Zeng-You, ZHENG Zhuo, ZHU Cheng. A fossil pollen dataset of China [J]. Chin J Plant Ecol, 2023, 47(10): 1453-1463. |
| [2] | CHEN Yu, NI Jian. QUANTITATIVE PALAEOVEGETATION RECONSTRUCTION AT LARGE SCALE BASED ON POLLEN RECORDS [J]. Chin J Plant Ecol, 2008, 32(5): 1201-1212. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Chinese Journal of Plant Ecology
Tel: 010-62836134, 62836138, E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn