

Chin J Plant Ecol ›› 2022, Vol. 46 ›› Issue (9): 1098-1108.DOI: 10.17521/cjpe.2022.0024
• Research Articles • Previous Articles Next Articles
ZHANG Hong-Xiang1,2,*( ), WEN Zhi-Bin1,2, WANG Qian1
), WEN Zhi-Bin1,2, WANG Qian1
Received:2022-01-13
Accepted:2022-03-09
Online:2022-09-20
Published:2022-10-19
Contact:
ZHANG Hong-Xiang
About author: E-mail: zhanghx561@ms.xjb.ac.cnSupported by:ZHANG Hong-Xiang, WEN Zhi-Bin, WANG Qian. Population genetic structure of Malus sieversii and environmental adaptations[J]. Chin J Plant Ecol, 2022, 46(9): 1098-1108.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.plant-ecology.com/EN/10.17521/cjpe.2022.0024
| 编号 Code | 来源地 Sampling site | 经度 Longitude (° E) | 纬度 Latitude (° N) | 观察杂合度 Observed heterozygosity | 期望杂合度 Expected heterozygosity |
|---|---|---|---|---|---|
| C1 | 中国新疆 Xinjiang, China | 83.61 | 43.38 | 0.328 4 (0.270 0) | 0.271 5 (0.174 3) |
| C2 | 中国新疆 Xinjiang, China | 82.86 | 43.26 | 0.316 5 (0.265 3) | 0.264 1 (0.177 1) |
| C3 | 中国新疆 Xinjiang, China | 81.62 | 44.12 | 0.332 0 (0.280 2) | 0.268 7 (0.179 3) |
| C4 | 中国新疆 Xinjiang, China | 80.78 | 44.43 | 0.329 0 (0.255 2) | 0.276 1 (0.168 5) |
| C5 | 哈萨克斯坦东部 Eastern Kazakhstan | 77.67 | 43.37 | 0.339 4 (0.244 8) | 0.288 2 (0.161 4) |
| C6 | 哈萨克斯坦东部 Eastern Kazakhstan | 77.68 | 43.36 | 0.337 6 (0.243 9) | 0.289 1 (0.160 1) |
| C7 | 吉尔吉斯斯坦 Kyrgyzstan | 73.62 | 41.90 | 0.346 5 (0.254 8) | 0.284 9 (0.165 9) |
| C8 | 吉尔吉斯斯坦 Kyrgyzstan | 72.96 | 41.36 | 0.332 6 (0.245 8) | 0.284 3 (0.165 2) |
| C9 | 哈萨克斯坦西部 Western Kazakhstan | 70.26 | 42.67 | 0.332 3 (0.246 2) | 0.286 5 (0.163 2) |
| C10 | 哈萨克斯坦西部 Western Kazakhstan | 69.95 | 41.94 | 0.326 7 (0.279 1) | 0.269 0 (0.180 4) |
Table 1 Code, sampling sites, geographical locations and genetic diversity of ten populations of Malus sieversii
| 编号 Code | 来源地 Sampling site | 经度 Longitude (° E) | 纬度 Latitude (° N) | 观察杂合度 Observed heterozygosity | 期望杂合度 Expected heterozygosity |
|---|---|---|---|---|---|
| C1 | 中国新疆 Xinjiang, China | 83.61 | 43.38 | 0.328 4 (0.270 0) | 0.271 5 (0.174 3) |
| C2 | 中国新疆 Xinjiang, China | 82.86 | 43.26 | 0.316 5 (0.265 3) | 0.264 1 (0.177 1) |
| C3 | 中国新疆 Xinjiang, China | 81.62 | 44.12 | 0.332 0 (0.280 2) | 0.268 7 (0.179 3) |
| C4 | 中国新疆 Xinjiang, China | 80.78 | 44.43 | 0.329 0 (0.255 2) | 0.276 1 (0.168 5) |
| C5 | 哈萨克斯坦东部 Eastern Kazakhstan | 77.67 | 43.37 | 0.339 4 (0.244 8) | 0.288 2 (0.161 4) |
| C6 | 哈萨克斯坦东部 Eastern Kazakhstan | 77.68 | 43.36 | 0.337 6 (0.243 9) | 0.289 1 (0.160 1) |
| C7 | 吉尔吉斯斯坦 Kyrgyzstan | 73.62 | 41.90 | 0.346 5 (0.254 8) | 0.284 9 (0.165 9) |
| C8 | 吉尔吉斯斯坦 Kyrgyzstan | 72.96 | 41.36 | 0.332 6 (0.245 8) | 0.284 3 (0.165 2) |
| C9 | 哈萨克斯坦西部 Western Kazakhstan | 70.26 | 42.67 | 0.332 3 (0.246 2) | 0.286 5 (0.163 2) |
| C10 | 哈萨克斯坦西部 Western Kazakhstan | 69.95 | 41.94 | 0.326 7 (0.279 1) | 0.269 0 (0.180 4) |
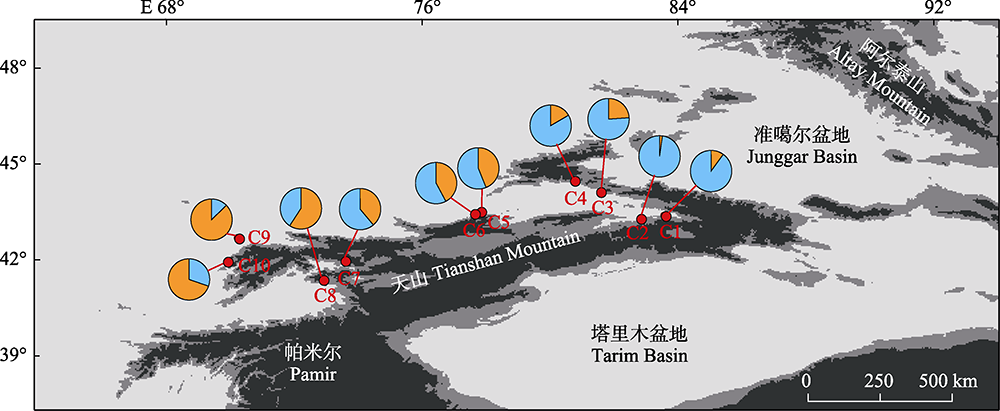
Fig. 1 Geographical locations and lineage distributions of ten Malus sieversii populations sampled. Pie charts show mean proportions of lineage memberships for each population from the ADMIXTURE program at K = 2; blue indicates lineage A, while orange indicates lineage B. Population codes are consistent with Table 1.
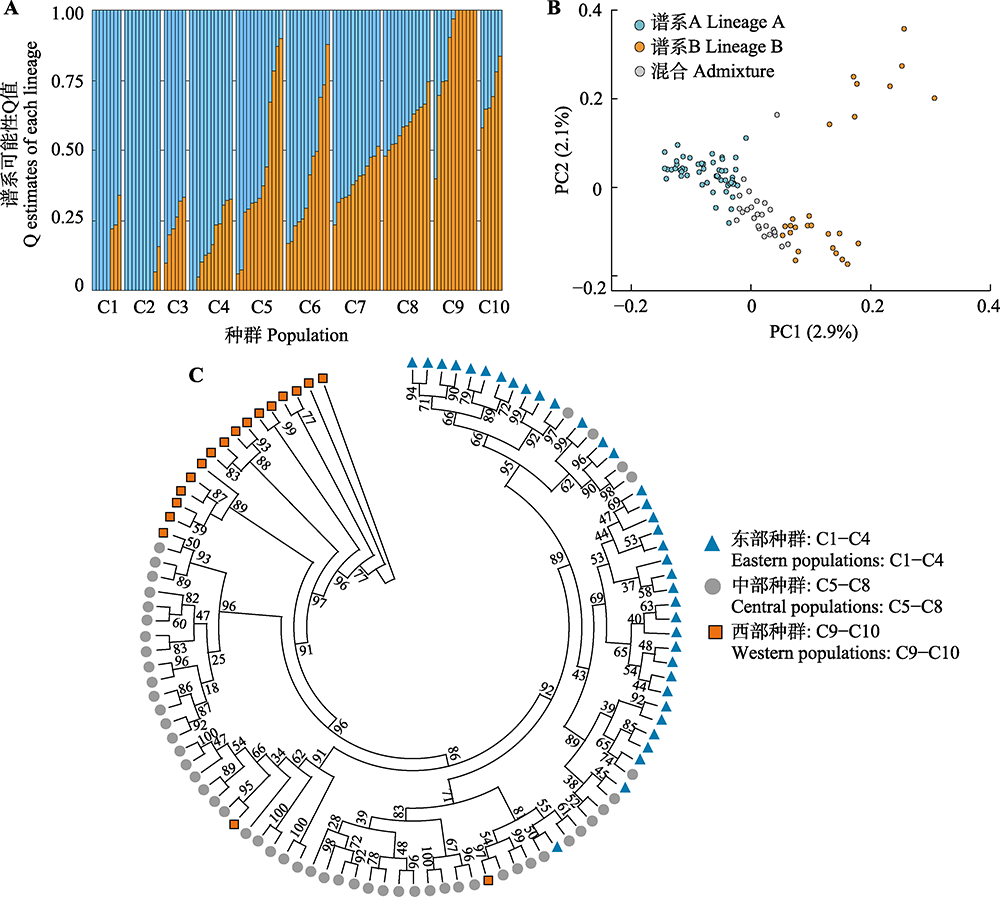
Fig. 2 Genetic clustering of the 105 individuals from ten populations of Malus sieversii. A, Result based on ADMIXTURE at K = 2. Blue columns indicate the Q estimates (possibility values) of lineage A, while orange columns indicate lineage B. B, Result based on principal component analysis (PCA). Individuals of each cluster were divided under the threshold of Q estimates (possibility values) ≥0.65, while individuals with Q estimates < 0.65 were defined as admixed. C, Result based on maximum-likelihood phylogenetic tree. Numbers near the branches showed the ultrafast bootstrap values of the nodes (%). Population codes are consistent with Table 1.
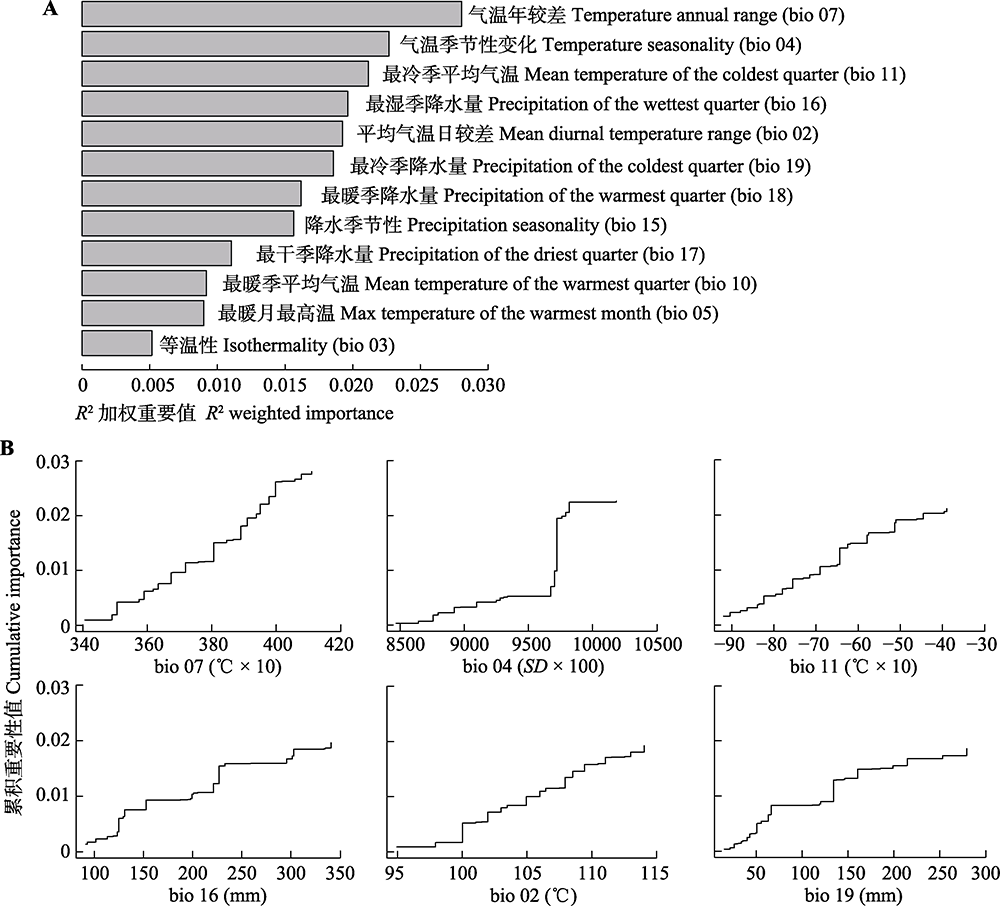
Fig. 3 Results of gradient forest (GF) analysis. A, R2-weighted importance of environmental variables that explain genetic gradients. B, Cumulative importance of allelic change along the first six environmental gradients.
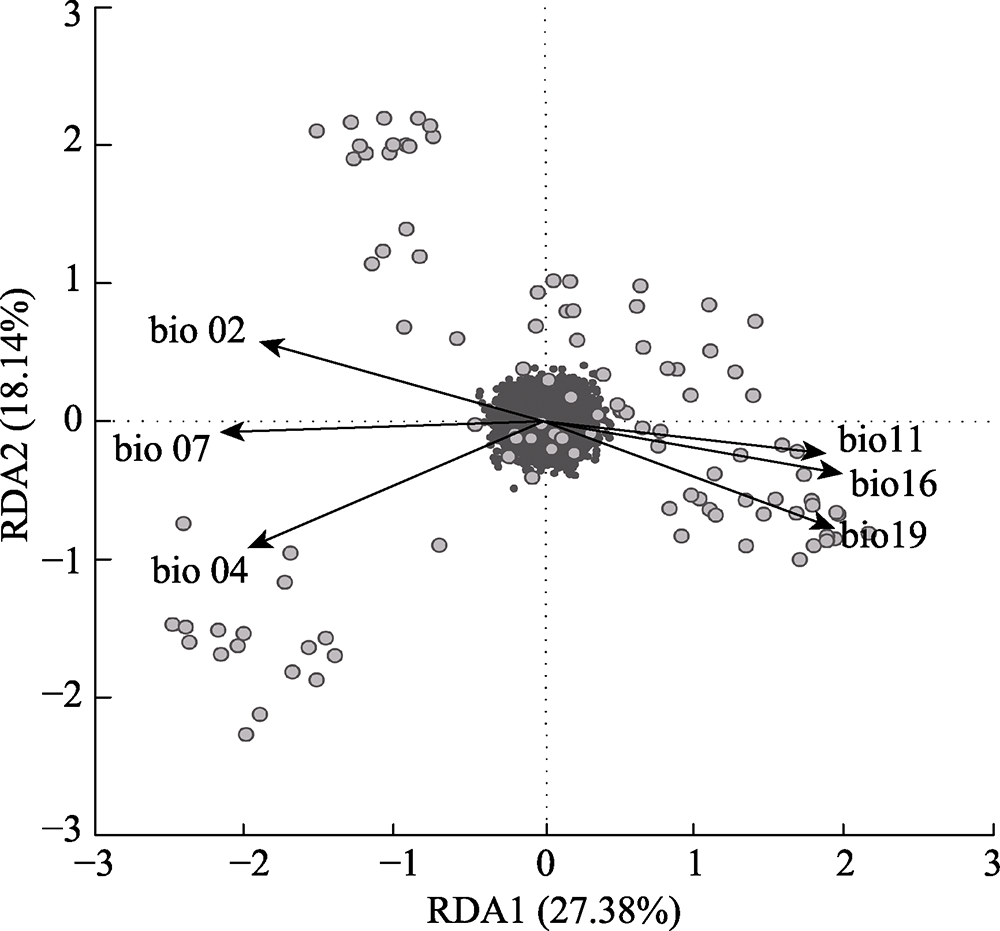
Fig. 4 Redundancy analysis (RDA) showing the relationship between environmental variables and genetic variations. Individuals are larger gray points. Small black points are single nucleotide polymorphism (SNP). bio 02, mean diurnal temperature range; bio 04, temperature seasonality; bio 07, temperature annual range; bio 11, mean temperature of the coldest quarter; bio 16, precipitation of the wettest quarter; bio 19, precipitation of the coldest quarter.
| 环境因子 Environmental variable | 约束比例 Constrained proportion | F | p |
|---|---|---|---|
| bio 07 | 0.019 3 | 2.031 | 0.001 |
| bio 04 | 0.018 6 | 1.951 | 0.001 |
| bio 11 | 0.017 4 | 1.827 | 0.001 |
| bio 16 | 0.017 9 | 1.883 | 0.001 |
| bio 02 | 0.017 7 | 1.860 | 0.001 |
| bio 19 | 0.018 0 | 1.887 | 0.001 |
Table 2 Redundancy analysis (RDA) based on six important environmental variables
| 环境因子 Environmental variable | 约束比例 Constrained proportion | F | p |
|---|---|---|---|
| bio 07 | 0.019 3 | 2.031 | 0.001 |
| bio 04 | 0.018 6 | 1.951 | 0.001 |
| bio 11 | 0.017 4 | 1.827 | 0.001 |
| bio 16 | 0.017 9 | 1.883 | 0.001 |
| bio 02 | 0.017 7 | 1.860 | 0.001 |
| bio 19 | 0.018 0 | 1.887 | 0.001 |
| 位点编号 Loci ID | 适应的气候因子 Adapted climate | 序列功能描述 Sequence description | 同源性E值 Blastn E-value |
|---|---|---|---|
| 1223* | bio 04 | 伴侣蛋白clpb1 Chaperone protein clpb1-like | 1.00E-101 |
| 2427* | bio 04, bio 07 | gdsl酯酶/脂肪酶at1g54790 x4亚型 gdsl esterase/lipase at1g54790-like isoform x4 | 1.00E-101 |
| 2666* | bio 04 | 自噬相关蛋白23 x2亚型 Autophagy-related protein 23-like isoform x2 | 1.00E-101 |
| 3144 | bio 04 | 12-氧代植二烯酸还原酶11 x2亚型 Putative 12-oxophytodienoate reductase 11 isoform x2 | 1.00E-101 |
| 3157* | bio 04 | 无义转录物upf3调控因子x6亚型 Regulator of nonsense transcripts upf3-like isoform x6 | 1.00E-101 |
| 4537 | bio 02, bio 04, bio 07 | 电压门控钾通道亚基 Probable voltage-gated potassium channel subunit beta | 1.00E-101 |
| 5799* | bio 11 | 核糖体生物合成蛋白brx1-2 Ribosome biogenesis protein brx1 homolog 2-like | 1.00E-101 |
| 5897* | bio 04 | 酰基辅酶A硫酯酶, 线粒体 Acyl-coenzyme a thioesterase 9, mitochondrial-like | 1.00E-101 |
| 6586 | bio 16 | 苯丙氨酸-tRNA连接酶α亚基, 细胞质 Phenylalanine-tRNA ligase alpha subunit, cytoplasmic | 1.00E-101 |
| 6766 | bio 04 | 2-异丙基苹果酸合酶 2-isopropylmalate synthase a-like | 1.00E-101 |
| 6936 | bio 16 | 三角状五肽重复蛋白at4g21065 Pentatricopeptide repeat-containing protein at4g21065 | 1.00E-101 |
| 8821 | bio 04 | 谷酰基tRNA转移酶亚基A, 叶绿体/线粒体x1亚型 Glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial-like isoform x1 | 1.00E-101 |
| 9378 | bio 04 | 转录因子myb3r-1 Transcription factor myb3r-1-like | 1.00E-101 |
| 9683 | bio 04 | 丝氨酸/苏氨酸蛋白激酶蛋白ccr4 Serine/threonine-protein kinase-like protein ccr4 | 1.00E-101 |
| 9684* | bio 04 | 磷脂酰肌醇磷酸酶sac1 x1亚型 Phosphoinositide phosphatase sac1 isoform x1 | 1.00E-101 |
Table 3 List of 15 candidate loci under selection based on LFMM analyses that alignment to the genome of Malus domestica
| 位点编号 Loci ID | 适应的气候因子 Adapted climate | 序列功能描述 Sequence description | 同源性E值 Blastn E-value |
|---|---|---|---|
| 1223* | bio 04 | 伴侣蛋白clpb1 Chaperone protein clpb1-like | 1.00E-101 |
| 2427* | bio 04, bio 07 | gdsl酯酶/脂肪酶at1g54790 x4亚型 gdsl esterase/lipase at1g54790-like isoform x4 | 1.00E-101 |
| 2666* | bio 04 | 自噬相关蛋白23 x2亚型 Autophagy-related protein 23-like isoform x2 | 1.00E-101 |
| 3144 | bio 04 | 12-氧代植二烯酸还原酶11 x2亚型 Putative 12-oxophytodienoate reductase 11 isoform x2 | 1.00E-101 |
| 3157* | bio 04 | 无义转录物upf3调控因子x6亚型 Regulator of nonsense transcripts upf3-like isoform x6 | 1.00E-101 |
| 4537 | bio 02, bio 04, bio 07 | 电压门控钾通道亚基 Probable voltage-gated potassium channel subunit beta | 1.00E-101 |
| 5799* | bio 11 | 核糖体生物合成蛋白brx1-2 Ribosome biogenesis protein brx1 homolog 2-like | 1.00E-101 |
| 5897* | bio 04 | 酰基辅酶A硫酯酶, 线粒体 Acyl-coenzyme a thioesterase 9, mitochondrial-like | 1.00E-101 |
| 6586 | bio 16 | 苯丙氨酸-tRNA连接酶α亚基, 细胞质 Phenylalanine-tRNA ligase alpha subunit, cytoplasmic | 1.00E-101 |
| 6766 | bio 04 | 2-异丙基苹果酸合酶 2-isopropylmalate synthase a-like | 1.00E-101 |
| 6936 | bio 16 | 三角状五肽重复蛋白at4g21065 Pentatricopeptide repeat-containing protein at4g21065 | 1.00E-101 |
| 8821 | bio 04 | 谷酰基tRNA转移酶亚基A, 叶绿体/线粒体x1亚型 Glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial-like isoform x1 | 1.00E-101 |
| 9378 | bio 04 | 转录因子myb3r-1 Transcription factor myb3r-1-like | 1.00E-101 |
| 9683 | bio 04 | 丝氨酸/苏氨酸蛋白激酶蛋白ccr4 Serine/threonine-protein kinase-like protein ccr4 | 1.00E-101 |
| 9684* | bio 04 | 磷脂酰肌醇磷酸酶sac1 x1亚型 Phosphoinositide phosphatase sac1 isoform x1 | 1.00E-101 |
| [1] |
Alexander DH, Novembre J, Lange K (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Research, 19, 1655-1664.
DOI PMID |
| [2] |
Bai WN, Yan PC, Zhang BW, Woeste KE, Lin K, Zhang DY (2018). Demographically idiosyncratic responses to climate change and rapid Pleistocene diversification of the walnut genus Juglans (Juglandaceae) revealed by whole- genome sequences. New Phytologist, 217, 1726-1736.
DOI URL |
| [3] |
Bay RA, Harrigan RJ, Underwood VL, Gibbs HL, Smith TB, Ruegg K (2018). Genomic signals of selection predict climate-driven population declines in a migratory bird. Science, 359, 83-86.
DOI URL |
| [4] |
Bian JX, Cui LC, Wang XY, Yang G, Huo FL, Ling HB, Chen LQ, She KJ, Du XH, Levi B, Levi AJ, Yan ZG, Nie XJ, Song WN (2020). Genomic and phenotypic divergence in wild barley driven by microgeographic adaptation. Advanced Science, 7, 2000709. DOI: 10.1002/advs.202000709.
DOI |
| [5] |
Brown JL, Hill DJ, Dolan AM, Carnaval AC, Haywood AM (2018). PaleoClim, high spatial resolution paleoclimate surfaces for global land areas. Scientific Data, 5, 180254. DOI: 10.1038/sdata.2018.254.
DOI |
| [6] |
Browne L, Wright JW, Fitz-Gibbon S, Gugger PF, Sork VL (2019). Adaptational lag to temperature in valley oak (Quercus lobata) can be mitigated by genome-informed assisted gene flow. Proceedings of the National Academy of Sciences of the United States of America, 116, 25179-25185.
DOI PMID |
| [7] |
Capblancq T, Luu K, Blum MGB, Bazin E (2018). Evaluation of redundancy analysis to identify signatures of local adaptation. Molecular Ecology Resources, 18, 1223-1233.
DOI PMID |
| [8] | Chang HS (1973). On the eco-geographical characters and the problems of classification of the wild fruit-tree forest in the Ili valley of Sinkiang. Acta Botanica Sinica, 15, 239-253. |
| [张新时 (1973). 伊犁野果林的生态地理特征和群落学问题. 植物学报, 15, 239-253.] | |
| [9] | Cui DF, Liao WB, Yang HJ, Zhang HD (2006). Studies on the floristic composition and genesis of the wild fruit forest in Tianshan Mountains in China. Forest Research, 19, 555-560. |
| [崔大方, 廖文波, 羊海军, 张宏达 (2006). 中国伊犁天山野果林区系表征地理成分及区系发生的研究. 林业科学研究, 19, 555-560.] | |
| [10] |
Daccord N, Celton JM, Linsmith G, Becker C, Choisne N, Schijlen E, van de Geest H, Bianco L, Micheletti D, Velasco R, di Pierro EA, Gouzy J, Rees DJG, Guérif P, Muranty H, et al. (2017). High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nature Genetics, 49, 1099-1106.
DOI PMID |
| [11] |
Duan NB, Bai Y, Sun HH, Wang N, Ma YM, Li MJ, Wang X, Jiao C, Legall N, Mao LY, Wan SB, Wang K, He TM, Feng SQ, Zhang ZY, et al. (2017). Genome re-sequencing reveals the history of apple and supports a two-stage model for fruit enlargement. Nature Communications, 8, 249. DOI: 10.1038/s41467-017-00336-7.
DOI |
| [12] |
Emerson KJ, Merz CR, Catchen JM, Hohenlohe PA, Cresko WA, Bradshaw WE, Holzapfel CM (2010). Resolving postglacial phylogeography using high-throughput sequencing. Proceedings of the National Academy of Sciences of the United States of America, 107, 16196-16200.
DOI PMID |
| [13] |
Frichot E, François O (2015). LEA: an R package for landscape and ecological association studies. Methods in Ecology and Evolution, 6, 925-929.
DOI URL |
| [14] | Fu LG, Jin JM (1992). China Plant Red Book—Rare and Endangered Plants. Science Press, Beijing. |
| [傅立国, 金鉴明 (1992). 中国植物红皮书——稀有濒危植物. 科学出版社, 北京.] | |
| [15] |
Gabay G, Faigenboim A, Dahan Y, Izhaki Y, Itkin M, Malitsky S, Elkind Y, Flaishman MA (2019). Transcriptome analysis and metabolic profiling reveal the key role of α-linolenic acid in dormancy regulation of European pear. Journal of Experimental Botany, 70, 1017-1031.
DOI PMID |
| [16] |
Geng DL, Lu LY, Yan MJ, Shen XX, Jiang LJ, Li HY, Wang LP, Yan Y, Xu JD, Li CY, Yu JT, Ma FW, Guan QM (2019). Physiological and transcriptomic analyses of roots from Malus sieversii under drought stress. Journal of Integrative Agriculture, 18, 1280-1294.
DOI URL |
| [17] |
Guerrero-Sánchez VM, Castillejo MÁ, López-Hidalgo C, Alconada AMM, Jorrín-Novo JV, Rey MD (2021). Changes in the transcript and protein profiles of Quercus ilex seedlings in response to drought stress. Journal of Proteomics, 243, 104263. DOI: 10.1016/j.jprot.2021.104263.
DOI |
| [18] |
Gugger PF, Liang CT, Sork VL, Hodgskiss P, Wright JW (2018). Applying landscape genomic tools to forest management and restoration of Hawaiian koa (Acacia koa) in a changing environment. Evolutionary Applications, 11, 231-242.
DOI PMID |
| [19] | Hou B, Xu Z (2005). Relationship of the occurences and evolutions of wild-fruit forests with climatic factors in the Tianshan Mountain. Acta Botanica Boreali-Occidentalia Sinica, 25, 2266-2271. |
| [侯博, 许正 (2005). 天山野果林的发生、演变与气候因素的关系. 西北植物学报, 25, 2266-2271.] | |
| [20] | Jeffries DL, Copp GH, Lawson Handley L, Olsén KH, Sayer CD, Hänfling B (2016). Comparing RADseq and microsatellites to infer complex phylogeographic patterns, an empirical perspective in the Crucian carp, Carassius carassius, L. Molecular Ecology, 25, 2997-3018. |
| [21] | Jia HX, Liu GJ, Li JB, Zhang J, Sun P, Zhao ST, Zhou X, Lu MZ, Hu JJ (2020). Genome resequencing reveals demographic history and genetic architecture of seed salinity tolerance in Populus euphratica. Journal of Experimental Botany, 71, 4308-4320. |
| [22] | Jiang XL, Gardner EM, Meng HH, Deng M, Xu GB (2019). Land bridges in the Pleistocene contributed to flora assembly on the continental islands of South China: insights from the evolutionary history of Quercus championii. Molecular Phylogenetics and Evolution, 132, 36-45. |
| [23] |
Lee U, Rioflorido I, Hong SW, Larkindale J, Waters ER, Vierling E (2007). The Arabidopsis ClpB/Hsp100 family of proteins: chaperones for stress and chloroplast development. The Plant Journal, 49, 115-127.
DOI URL |
| [24] |
Li H, Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754-1760.
DOI PMID |
| [25] |
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R,1000 Genome Project Data Proc (2009). The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078-2079.
DOI PMID |
| [26] |
Li LF, Cushman SA, He YX, Ma XF, Ge XJ, Li JX, Qian ZH, Li Y (2022). Landscape genomics reveals genetic evidence of local adaptation in a widespread tree, the Chinese wingnut (Pterocarya stenoptera). Journal of Systematics and Evolution, 60, 386-397.
DOI URL |
| [27] |
Li WF, Mao J, Su J, Li XW, Dawuda MM, Ma ZH, Zuo CW, An ZS, Chen BH (2021). Exogenous ABA and its inhibitor regulate flower bud induction of apple cv. ‘Nagafu No. 2’ grafted on different rootstocks. Trees, 35, 609-620.
DOI URL |
| [28] | Lin PJ, Cui NR (2000). Wild Fruit Forest Resources in the Tianshan Mountains: a Comprehensive Research of Ili Wild Fruit Forest. China Forestry Publishing House, Beijing. |
| [林培钧, 崔乃然 (2000). 天山野果林资源——伊犁野果林综合研究. 中国林业出版社, 北京.] | |
| [29] | Liu JX, Qu CZ, Xu HF, Su MY, Fang HC, Wang N, Jiang SH, Wang YC, Zhang ZY, Chen XS (2017). Evaluation on fruit flavor quality in two second-generation hybrid apple lines. Journal of Fruit Science, 34, 988-995. |
| [刘静轩, 曲常志, 许海峰, 苏梦雨, 房鸿成, 王楠, 姜生辉, 王意程, 张宗营, 陈学森 (2017). 新疆红肉苹果杂交二代2个功能型株系果实风味品质的评价. 果树学报, 34, 988-995.] | |
| [30] | Liu XS, Lin PJ, Zhong JP (1993). An analysis and inquiry into the wild apple trees in Iri. Arid Zone Research, 10(3), 28-33. |
| [刘兴诗, 林培钧, 钟骏平 (1993). 伊犁野果林生境分析和发生探讨. 干旱区研究, 10(3), 28-33.] | |
| [31] |
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research, 20, 1297-1303.
DOI PMID |
| [32] |
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating Maximum-Likelihood phylogenies. Molecular Biology and Evolution, 32, 268-274.
DOI URL |
| [33] | Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens HH, Szoecs E, Wagner H (2018). Vegan v2.6: community ecology package. [2019-09-01]. https://CRAN.R-project.org/package=vegan. |
| [34] |
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, Daly MJ, Sham PC (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics, 81, 559-575.
DOI URL |
| [35] |
Tani T, Sobajima H, Okada K, Chujo T, Arimura SI, Tsutsumi N, Nishimura M, Seto H, Nojiri H, Yamane H (2008). Identification of the OsOPR7 gene encoding 12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta, 227, 517-526.
DOI PMID |
| [36] |
Tao Y, Zhang YM, Zhou XB (2016). Ecological stoichiometry of surface soil nutrient and its influencing factors in the wild fruit forest in Yili region, Xinjiang, China. Chinese Journal of Applied Ecology, 27, 2239-2248.
DOI |
|
[陶冶, 张元明, 周晓兵 (2016). 伊犁野果林浅层土壤养分生态化学计量特征及其影响因素. 应用生态学报, 27, 2239-2248.]
DOI |
|
| [37] |
Tian ZP, Zhuang L, Li JG, Cheng MX (2011a). Interspecific and environmental relationships of woody plant species in wild fruit-tree forests on the north slope of Ili Valley. Biodiversity Science, 19, 335-342.
DOI URL |
| [田中平, 庄丽, 李建贵, 程模香 (2011a). 伊犁河谷北坡野果林木本植物种间关系及环境解释. 生物多样性, 19, 335-342.] | |
| [38] | Tian ZP, Zhuang L, Li JG, Xu ZQ, Zhang L (2011b). Relationship between community structure of wild fruit forests and their environment on north-facing slopes of the Iri Valley. Chinese Journal of Applied & Environmental Biology, 17, 39-45. |
| [田中平, 庄丽, 李建贵, 徐智全, 张莉 (2011b). 伊犁河谷北坡野果林群落结构及其与环境的关系. 应用与环境生物学报, 17, 39-45.] | |
| [39] |
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, Salvi S, Pindo M, Baldi P, Castelletti S, Cavaiuolo M, et al. (2010). The genome of the domesticated apple (Malus × domestica Borkh.). Nature Genetics, 42, 833-839.
DOI |
| [40] | Wang J, Liao K, Zhao L, Wang YL (2009). Identification of cold-resistance in Prunus divaricata. Xinjiang Agricultural Sciences, 46, 289-292. |
| [王瑾, 廖康, 赵蕾, 王燕凌 (2009). 野生樱桃李枝条抗寒性鉴定. 新疆农业科学, 46, 289-292.] | |
| [41] |
Wang N, Jiang S, Zhang Z, Fang H, Xu H, Wang Y, Chen X (2018). Malus sieversii: the origin, flavonoid synthesis mechanism, and breeding of red-skinned and red-fleshed apples. Horticulture Research, 5, 70. DOI: 10.1038/s41438-018-0084-4.
DOI |
| [42] |
Wang YT, Feng C, Zhai ZF, Peng X, Wang YY, Sun YT, Li J, Shen XS, Xiao YQ, Zhu SJ, Huang XW, Li TH (2020). The apple microR171i-SCARECROW-LIKE PROTEINS26.1 module enhances drought stress tolerance by integrating ascorbic acid metabolism. Plant Physiology, 184, 194-211.
DOI URL |
| [43] |
Xie ZZ, Barski OA, Cai J, Bhatnagar A, Tipparaju SM (2011). Catalytic reduction of carbonyl groups in oxidized PAPC by Kvβ2 (AKR6). Chemico-Biological Interactions, 191, 255-260.
DOI PMID |
| [44] | Xu JN, Liu G, Ma P, Huang YP, Yang YL (2014). Research progress of adversity response mechanism of Malus sieversii. Shandong Agricultural Sciences, 46(8), 138-141. |
| [徐佳宁, 刘钢, 马盼, 黄翊鹏, 杨艳丽 (2014). 新疆野苹果逆境响应机制研究进展. 山东农业科学, 46(8), 138-141.] | |
| [45] |
Yang J, Lee SH, Goddard ME, Visscher PM (2011). GCTA: a tool for genome-wide complex trait analysis. The American Journal of Human Genetics, 88, 76-82.
DOI URL |
| [46] |
Yang ML, Che SY, Zhang YX, Wang HB, Wei T, Yan GR, Song WQ, Yu WW (2019). Universal stress protein in Malus sieversii confers enhanced drought tolerance. Journal of Plant Research, 132, 825-837.
DOI URL |
| [47] | Zhang B, Liu LQ, Qin W, Wurenqimige (2021). Study on physiological and biochemical mechanism of cold resistance of Malus sieversii Non-Wood Forest Research, 39, 60-68. |
| [张博, 刘立强, 秦伟, 乌仁其米格 (2021). 新疆野苹果抗寒生理生化机制研究. 经济林研究, 39, 60-68.] | |
| [48] |
Zhang HX, Li XS, Wang JC, Zhang DY (2021). Insights into the aridification history of Central Asian Mountains and international conservation strategy from the endangered wild apple tree. Journal of Biogeography, 48, 332-344.
DOI URL |
| [49] |
Zhang HX, Zhang ML, Wang LN (2015). Genetic structure and historical demography of Malus sieversii in the Yili Valley and the western mountains of the Junggar Basin, Xinjiang, China. Journal of Arid Land, 7, 264-271.
DOI URL |
| [50] | Zhang HX, Zheng TY (2020a). Effect of habitat fragmentation on the population genetic structure of Malus sieversii. Arid Zone Research, 37, 715-721. |
| [张宏祥, 郑田勇 (2020a). 生境片段化对新疆野苹果种群遗传结构的影响. 干旱区研究, 37, 715-721.] | |
| [51] | Zhang HX, Zheng TY (2020b). Effects of elevation on population genetic characteristics of Malus sieversii. Chinese Journal of Ecology, 39, 4031-4037. |
| [张宏祥, 郑田勇 (2020b). 海拔对新疆野苹果种群遗传特征的影响. 生态学杂志, 39, 4031-4037.] | |
| [52] | Zhang MH, Wang JC, Yang HL, Zhang DY, Deng PC, Cui ZJ (2020). Physiological response of Xinjiang wild walnut germplasm to low temperature stress. Chinese Journal of Applied Ecology, 31, 2558-2566. |
|
[张敏欢, 王建成, 杨红兰, 张道远, 邓鹏程, 崔志军 (2020). 新疆野核桃种质对低温胁迫的生理响应. 应用生态学报, 31, 2558-2566.]
DOI |
|
| [53] |
Zhang Z, Schwartz S, Wagner L, Miller W (2000). A greedy algorithm for aligning DNA sequences. Journal of Computational Biology, 7, 203-214.
PMID |
| [54] |
Zhao YP, Fan G, Yin PP, Sun S, Li N, Hong XN, Hu G, Zhang H, Zhang FM, Han JD, Hao YJ, Xu Q, Yang X, Xia W, Chen W, et al. (2019). Resequencing 545 ginkgo genomes across the world reveals the evolutionary history of the living fossil. Nature Communications, 10, 4201. DOI: 10.1038/s41467-019-12133-5.
DOI |
| [55] | Zheng D, Wu YX, Qin WM, He TM (2019). Advance in research on application of Malus sieversii as rootstock. Chinese Wild Plant Resources, 38(2), 56-59. |
| [郑点, 吴玉霞, 覃伟铭, 何天明 (2019). 新疆野苹果作为苹果砧木利用的研究进展. 中国野生植物资源, 38(2), 56-59.] | |
| [56] | Zhong HX, Lu CS, Luo SP, Li J (2016). The study of cold resistance test of dormancy branches and buds of Amygdalus ledebouriana schleche in Xinjiang. Xinjiang Agricultural Sciences, 53, 120-125. |
| [钟海霞, 卢春生, 罗淑萍, 李疆 (2016). 新疆野扁桃休眠枝条和花芽的抗寒试验研究. 新疆农业科学, 53, 120-125.] | |
| [57] |
Zhou P, Li XS, Liu XJ, Wen XJ, Zhang Y, Zhang DY (2021). Transcriptome profiling of Malus sieversii under freezing stress after being cold-acclimated. BMC Genomics, 22, 681. DOI: 10.1186/s12864-021-07998-0.
DOI |
| [1] | HE Lu-Lu, ZHANG Xuan, ZHANG Yu-Wen, WANG Xiao-Xia, LIU Ya-Dong, LIU Yan, FAN Zi-Ying, HE Yuan-Yang, XI Ben-Ye, DUAN Jie. Crown characteristics and its relationship with tree growth on different slope aspects for Larix olgensis var. changbaiensis plantation in eastern Liaoning mountainous area, China [J]. Chin J Plant Ecol, 2023, 47(11): 1523-1539. |
| [2] | WANG Chun-Cheng, MA Song-Mei, ZHANG Dan, WANG Shao-Ming. Spatial genetic structure of Lycium ruthenicum in the Qaidam Basin [J]. Chin J Plant Ecol, 2020, 44(6): 661-668. |
| [3] | CHAI Yong-Fu, XU Jin-Shi, LIU Hong-Yan, LIU Quan-Ru, ZHENG Cheng-Yang, KANG Mu-Yi, LIANG Cun-Zhu, WANG Ren-Qing, GAO Xian-Ming, ZHANG Feng, SHI Fu-Chen, LIU Xiao, YUE Ming. Species composition and phylogenetic structure of major shrublands in North China [J]. Chin J Plant Ecol, 2019, 43(9): 793-805. |
| [4] | Ming-Fei ZHAO, Feng XUE, Yu-Hang WANG, Guo-Yi WANG, Kai-Xiong XING, Mu-Yi KANG, Jing-Lan WANG. Phylogenetic structure and diversity of herbaceous communities in the conifer forests along an elevational gradient in Luya Mountain, Shanxi, China [J]. Chin J Plan Ecolo, 2017, 41(7): 707-715. |
| [5] | HUANG Jian-Xiong, ZHENG Feng-Ying, MI Xiang-Cheng. Influence of environmental factors on phylogenetic structure at multiple spatial scales in an evergreen broad-leaved forest of China [J]. Chin J Plant Ecol, 2010, 34(3): 309-315. |
| [6] | YAN Bang-Guo, WEN Wei-Quan, ZHANG Jian, YANG Wan-Qin, LIU Yang, HUANG Xu, LI Ze-Bo. Plant community assembly rules across a subalpine grazing gradient in western Sichuan, China [J]. Chin J Plant Ecol, 2010, 34(11): 1294-1302. |
| [7] | CHEN Liang-Hua, HU Ting-Xing, ZHANG Fan, LI Guo-He. GENETIC DIVERSITIES OF FOUR JUGLANSPOPULATIONS REVEALED BY AFLP IN SICHUAN PROVINCE, CHINA [J]. Chin J Plant Ecol, 2008, 32(6): 1362-1372. |
| [8] | WANG Ying, KANG Ming, HUANG Hong-Wen. SUBPOPULATION GENETIC STRUCTURE IN A PANMICTIC POPULATION AS REVEALED BY MOLECULAR MARKERS: A CASE STUDY OF CASTANEA SEQUINII USING SSR MARKERS [J]. Chin J Plant Ecol, 2006, 30(1): 147-156. |
| [9] | CHEN Xiao-Yong. Spatial Autocorrelation of Genetic Structure in a Population of Cyclobalanopsis glauca in Huangshan, Anhui [J]. Chin J Plan Ecolo, 2001, 25(1): 29-34. |
| [10] | Zheng Huiying, Shen Quanguan, Yan Tian. The Ecological and Physiological Adaptation of Kochia sieversiana and the Characteristics of the Communities [J]. Chin J Plan Ecolo, 1998, 22(1): 1-7. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2026 Chinese Journal of Plant Ecology
Tel: 010-62836134, 62836138, E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
![]()