

植物生态学报 ›› 2012, Vol. 36 ›› Issue (6): 560-571.DOI: 10.3724/SP.J.1258.2012.00560
彭兴民1,2,*( ), 吴疆翀1, 郑益兴1, 张燕平1, 李根前2
), 吴疆翀1, 郑益兴1, 张燕平1, 李根前2
收稿日期:2012-02-13
接受日期:2012-04-09
出版日期:2012-02-13
发布日期:2012-06-04
通讯作者:
彭兴民
作者简介:*(E-mail:jshe@pku.edu.cn)
PENG Xing-Min1,2,*( ), WU Jiang-Chong1, ZHENG Yi-Xing1, ZHANG Yan-Ping1, LI Gen-Qian2
), WU Jiang-Chong1, ZHENG Yi-Xing1, ZHANG Yan-Ping1, LI Gen-Qian2
Received:2012-02-13
Accepted:2012-04-09
Online:2012-02-13
Published:2012-06-04
Contact:
PENG Xing-Min
摘要:
为了揭示印楝(Azadirachta indica)实生种群表型变异程度和变异规律, 以云南引种印楝人工林为研究对象, 基于9个种群90个单株14个表型性状严格细致的测量, 采用单因素方差分析、巢式方差分析、相关分析、协方差主成分分析(S法)和非加权配对算术平均法(UPGMA)聚类分析等数理方法, 分析了种群的表型变异。结果表明: 印楝种内表型性状在种群间和种群内均存在着较丰富的差异, 种群内的变异大于种群间的变异, 种群间的分化相对较小。对表型性状进行的变异系数多重比较和协方差主成分分析(S法)均显示, 结实和种子化学成分相关性状的变异是造成印楝表型变异的主要来源。利用种群间欧氏距离进行的UPGMA聚类分析结果进一步表明, 印楝9个种群可以分为4类, 表型性状并没有严格依地理距离而聚类。研究结果为印楝的遗传改良工作奠定了基础, 为制定育种策略和人工经营对策提供了科学依据。
彭兴民, 吴疆翀, 郑益兴, 张燕平, 李根前. 云南引种印楝实生种群的表型变异. 植物生态学报, 2012, 36(6): 560-571. DOI: 10.3724/SP.J.1258.2012.00560
PENG Xing-Min, WU Jiang-Chong, ZHENG Yi-Xing, ZHANG Yan-Ping, LI Gen-Qian. Phenotypic variation in cultivated populations of Azadirachta indica in Yunnan, China. Chinese Journal of Plant Ecology, 2012, 36(6): 560-571. DOI: 10.3724/SP.J.1258.2012.00560
| 种群编号及农业 生态气候区 Population code and agro-ecological region | 种群 Population | 纬度 Latitude (N) | 经度 Longitude (E) | 海拔 Elevation (m) | 年平均气温 Annual mean air temperature (℃) | 年降水量 Annual precipitation (mm) | 生境 Habitat |
|---|---|---|---|---|---|---|---|
| P1a | XBL | 25°48′ | 101°51′ | 1 135.0 | 21.7 | 645.0 | 农田旁 Cultivated land side |
| P2a | WX | 25°44′ | 101°52′ | 1 120.2 | 21.8 | 634.0 | 新垦台地 Land reclaimed |
| P3a | NMS | 25°37′ | 101°53′ | 1 277.0 | 20.9 | 645.0 | 较陡荒坡 Steep Hillside |
| P4b | DHG | 23°19′ | 102°34′ | 553.0 | 22.8 | 843.0 | 平缓荒坡 Gradual hillside |
| P5b | WW | 23°18′ | 102°38′ | 430.0 | 23.5 | 840.0 | 平缓荒坡 Gradual hillside |
| P6b | NS | 23°13′ | 102°50′ | 257.1 | 24.5 | 945.8 | 耕地 Cultivated land |
| P7b | LD | 23°12′ | 102°55′ | 223.0 | 24.7 | 945.8 | 洪积台地 Flood land |
| P8b | MT | 23°08′ | 103°03′ | 197.0 | 24.8 | 980.0 | 平缓荒坡 Gradual hillside |
| P9b | HCB | 23°06′ | 103°11′ | 256.0 | 24.5 | 1 060.0 | 平缓荒坡 Gradual hillside |
表1 印楝取样种群的概况
Table 1 Information of sampled populations of Azadirachta indica
| 种群编号及农业 生态气候区 Population code and agro-ecological region | 种群 Population | 纬度 Latitude (N) | 经度 Longitude (E) | 海拔 Elevation (m) | 年平均气温 Annual mean air temperature (℃) | 年降水量 Annual precipitation (mm) | 生境 Habitat |
|---|---|---|---|---|---|---|---|
| P1a | XBL | 25°48′ | 101°51′ | 1 135.0 | 21.7 | 645.0 | 农田旁 Cultivated land side |
| P2a | WX | 25°44′ | 101°52′ | 1 120.2 | 21.8 | 634.0 | 新垦台地 Land reclaimed |
| P3a | NMS | 25°37′ | 101°53′ | 1 277.0 | 20.9 | 645.0 | 较陡荒坡 Steep Hillside |
| P4b | DHG | 23°19′ | 102°34′ | 553.0 | 22.8 | 843.0 | 平缓荒坡 Gradual hillside |
| P5b | WW | 23°18′ | 102°38′ | 430.0 | 23.5 | 840.0 | 平缓荒坡 Gradual hillside |
| P6b | NS | 23°13′ | 102°50′ | 257.1 | 24.5 | 945.8 | 耕地 Cultivated land |
| P7b | LD | 23°12′ | 102°55′ | 223.0 | 24.7 | 945.8 | 洪积台地 Flood land |
| P8b | MT | 23°08′ | 103°03′ | 197.0 | 24.8 | 980.0 | 平缓荒坡 Gradual hillside |
| P9b | HCB | 23°06′ | 103°11′ | 256.0 | 24.5 | 1 060.0 | 平缓荒坡 Gradual hillside |
| 种群 Population | SL (cm) | SW (cm) | SLW | ST (g) | KT (g) | SKT (%) | SG (m) |
|---|---|---|---|---|---|---|---|
| XBL | 1.37 ± 0.15abc | 0.72 ± 0.08a | 1.95 ± 0.37 | 0.19 ± 0.03b | 0.10 ± 0.02bc | 51.30 ± 1.12 | 6.47 ± 1.45cd |
| WX | 1.31 ± 0.15c | 0.65 ± 0.07b | 2.05 ± 0.38 | 0.18 ± 0.02b | 0.09 ± 0.02c | 51.10 ± 1.53 | 5.54 ± 1.08d |
| NMS | 1.33 ± 0.16bc | 0.69 ± 0.08ab | 1.98 ± 0.37 | 0.19 ± 0.03b | 0.10 ± 0.02bc | 51.12 ± 0.85 | 4.07 ± 0.63e |
| DHG | 1.31 ± 0.17c | 0.71 ± 0.09ab | 1.91 ± 0.39 | 0.19 ± 0.04b | 0.10 ± 0.02bc | 51.13 ± 1.29 | 6.93 ± 0.89bc |
| WW | 1.43 ± 0.19ab | 0.69 ± 0.07ab | 2.11 ± 0.37 | 0.21 ± 0.04ab | 0.11 ± 0.02abc | 51.29 ± 1.28 | 6.75 ± 1.43bc |
| NS | 1.45 ± 0.17a | 0.72 ± 0.08a | 2.04 ± 0.41 | 0.22 ± 0.04ab | 0.11 ± 0.02ab | 51.83 ± 2.29 | 7.71 ± 1.32ab |
| LD | 1.43 ± 0.18ab | 0.69 ± 0.08ab | 2.09 ± 0.42 | 0.24 ± 0.04a | 0.12 ± 0.02a | 51.12 ± 2.50 | 8.11 ± 1.18a |
| MT | 1.42 ± 0.19ab | 0.66 ± 0.07b | 2.20 ± 0.40 | 0.21 ± 0.04ab | 0.11 ± 0.02abc | 51.57 ± 2.37 | 7.61 ± 0.78ab |
| HCB | 1.38 ± 0.19abc | 0.70 ± 0.08ab | 1.98 ± 0.40 | 0.21 ± 0.04ab | 0.11 ± 0.02abc | 51.60 ± 2.33 | 5.64 ± 1.07d |
| 平均值 Average | 1.38 ± 0.17 | 0.69 ± 0.08 | 2.04 ± 0.49 | 0.21 ± 0.04 | 0.11 ± 0.02 | 51.34 ± 1.73 | 6.54 ± 1.62 |
| 种群间F值 F value among populations | 2.60* | 2.22* | 1.38 | 2.34* | 3.85** | 1.24 | 12.90** |
| 种群内F值 F value within population | 9.11** | 6.57** | 5.73** | 20.50** | 14.40** | 3.33** | - |
| 种群 Population | ZG (m) | DJ (cm) | GF (m) | GS (grain) | azA (%) | azB (%) | azAB (%) |
| XBL | 0.58 ± 0.50cd | 17.06 ± 2.92ab | 6.50 ± 1.74bcd | 4240 ± 23.20b | 0.50 ± 5.21cde | 0.16 ± 3.39b | 0.66 ± 8.41cd |
| WX | 0.38 ± 0.32cd | 14.17 ± 3.76bc | 4.86 ± 1.62de | 3900 ± 16.21b | 0.49 ± 6.44de | 0.16 ± 2.37b | 0.65 ± 7.90cd |
| NMS | 0.40 ± 0.38cd | 12.38 ± 1.35c | 4.25 ± 0.94e | 1990 ± 11.64c | 0.56 ± 4.59abcd | 0.20 ± 2.19a | 0.77 ± 7.58ab |
| DHG | 0.35 ± 0.34d | 17.55 ± 4.33ab | 8.24 ± 1.74ab | 6020 ± 14.86ab | 0.59 ± 3.06ab | 0.19 ± 2.78a | 0.79 ± 5.15ab |
| WW | 0.81 ± 0.31bc | 16.45 ± 5.12ab | 8.46 ± 2.42a | 4426 ± 17.49b | 0.57 ± 2.57abc | 0.16 ± 2.11b | 0.73 ± 3.13bc |
| NS | 1.28 ± 0.47a | 18.24 ± 5.62ab | 8.26 ± 2.09ab | 5360 ± 25.26ab | 0.47 ± 3.73e | 0.15 ± 2.29b | 0.63 ± 4.74d |
| LD | 1.27 ± 0.65a | 19.96 ± 4.02a | 7.78 ± 1.95ab | 6050 ± 14.46ab | 0.62 ± 6.05a | 0.21 ± 1.89a | 0.83 ± 8.81a |
| MT | 1.03 ± 0.48ab | 15.03 ± 4.10bc | 6.03 ± 1.84cd | 7160 ± 20.39a | 0.53 ± 2.92bcde | 0.11 ± 2.26c | 0.65 ± 2.84cd |
| HCB | 0.46 ± 0.39cd | 17.28 ± 3.92ab | 6.80 ± 1.28abc | 3824 ± 10.63b | 0.53 ± 4.13bcde | 0.14 ± 2.99bc | 0.67 ± 5.40cd |
| 平均值 Average | 0.73 ± 0.56 | 16.46 ± 4.45 | 6.79 ± 2.24 | 4774 ± 20.23 | 0.54 ± 5.06 | 0.17 ± 3.38 | 0.71 ± 7.74 |
| 种群间F值 F value among populations | 7.36** | 3.13** | 7.41** | 4.19** | 4.01** | 10.07** | 6.38** |
| 种群内F值 F value within population | - | - | - | - | - | - | - |
表2 印楝9个种群表型性状的方差分析(平均值±标准偏差)
Table 2 Variance analysis of phenotypic traits variance between nine Azadirachta indica populations (mean ± SD)
| 种群 Population | SL (cm) | SW (cm) | SLW | ST (g) | KT (g) | SKT (%) | SG (m) |
|---|---|---|---|---|---|---|---|
| XBL | 1.37 ± 0.15abc | 0.72 ± 0.08a | 1.95 ± 0.37 | 0.19 ± 0.03b | 0.10 ± 0.02bc | 51.30 ± 1.12 | 6.47 ± 1.45cd |
| WX | 1.31 ± 0.15c | 0.65 ± 0.07b | 2.05 ± 0.38 | 0.18 ± 0.02b | 0.09 ± 0.02c | 51.10 ± 1.53 | 5.54 ± 1.08d |
| NMS | 1.33 ± 0.16bc | 0.69 ± 0.08ab | 1.98 ± 0.37 | 0.19 ± 0.03b | 0.10 ± 0.02bc | 51.12 ± 0.85 | 4.07 ± 0.63e |
| DHG | 1.31 ± 0.17c | 0.71 ± 0.09ab | 1.91 ± 0.39 | 0.19 ± 0.04b | 0.10 ± 0.02bc | 51.13 ± 1.29 | 6.93 ± 0.89bc |
| WW | 1.43 ± 0.19ab | 0.69 ± 0.07ab | 2.11 ± 0.37 | 0.21 ± 0.04ab | 0.11 ± 0.02abc | 51.29 ± 1.28 | 6.75 ± 1.43bc |
| NS | 1.45 ± 0.17a | 0.72 ± 0.08a | 2.04 ± 0.41 | 0.22 ± 0.04ab | 0.11 ± 0.02ab | 51.83 ± 2.29 | 7.71 ± 1.32ab |
| LD | 1.43 ± 0.18ab | 0.69 ± 0.08ab | 2.09 ± 0.42 | 0.24 ± 0.04a | 0.12 ± 0.02a | 51.12 ± 2.50 | 8.11 ± 1.18a |
| MT | 1.42 ± 0.19ab | 0.66 ± 0.07b | 2.20 ± 0.40 | 0.21 ± 0.04ab | 0.11 ± 0.02abc | 51.57 ± 2.37 | 7.61 ± 0.78ab |
| HCB | 1.38 ± 0.19abc | 0.70 ± 0.08ab | 1.98 ± 0.40 | 0.21 ± 0.04ab | 0.11 ± 0.02abc | 51.60 ± 2.33 | 5.64 ± 1.07d |
| 平均值 Average | 1.38 ± 0.17 | 0.69 ± 0.08 | 2.04 ± 0.49 | 0.21 ± 0.04 | 0.11 ± 0.02 | 51.34 ± 1.73 | 6.54 ± 1.62 |
| 种群间F值 F value among populations | 2.60* | 2.22* | 1.38 | 2.34* | 3.85** | 1.24 | 12.90** |
| 种群内F值 F value within population | 9.11** | 6.57** | 5.73** | 20.50** | 14.40** | 3.33** | - |
| 种群 Population | ZG (m) | DJ (cm) | GF (m) | GS (grain) | azA (%) | azB (%) | azAB (%) |
| XBL | 0.58 ± 0.50cd | 17.06 ± 2.92ab | 6.50 ± 1.74bcd | 4240 ± 23.20b | 0.50 ± 5.21cde | 0.16 ± 3.39b | 0.66 ± 8.41cd |
| WX | 0.38 ± 0.32cd | 14.17 ± 3.76bc | 4.86 ± 1.62de | 3900 ± 16.21b | 0.49 ± 6.44de | 0.16 ± 2.37b | 0.65 ± 7.90cd |
| NMS | 0.40 ± 0.38cd | 12.38 ± 1.35c | 4.25 ± 0.94e | 1990 ± 11.64c | 0.56 ± 4.59abcd | 0.20 ± 2.19a | 0.77 ± 7.58ab |
| DHG | 0.35 ± 0.34d | 17.55 ± 4.33ab | 8.24 ± 1.74ab | 6020 ± 14.86ab | 0.59 ± 3.06ab | 0.19 ± 2.78a | 0.79 ± 5.15ab |
| WW | 0.81 ± 0.31bc | 16.45 ± 5.12ab | 8.46 ± 2.42a | 4426 ± 17.49b | 0.57 ± 2.57abc | 0.16 ± 2.11b | 0.73 ± 3.13bc |
| NS | 1.28 ± 0.47a | 18.24 ± 5.62ab | 8.26 ± 2.09ab | 5360 ± 25.26ab | 0.47 ± 3.73e | 0.15 ± 2.29b | 0.63 ± 4.74d |
| LD | 1.27 ± 0.65a | 19.96 ± 4.02a | 7.78 ± 1.95ab | 6050 ± 14.46ab | 0.62 ± 6.05a | 0.21 ± 1.89a | 0.83 ± 8.81a |
| MT | 1.03 ± 0.48ab | 15.03 ± 4.10bc | 6.03 ± 1.84cd | 7160 ± 20.39a | 0.53 ± 2.92bcde | 0.11 ± 2.26c | 0.65 ± 2.84cd |
| HCB | 0.46 ± 0.39cd | 17.28 ± 3.92ab | 6.80 ± 1.28abc | 3824 ± 10.63b | 0.53 ± 4.13bcde | 0.14 ± 2.99bc | 0.67 ± 5.40cd |
| 平均值 Average | 0.73 ± 0.56 | 16.46 ± 4.45 | 6.79 ± 2.24 | 4774 ± 20.23 | 0.54 ± 5.06 | 0.17 ± 3.38 | 0.71 ± 7.74 |
| 种群间F值 F value among populations | 7.36** | 3.13** | 7.41** | 4.19** | 4.01** | 10.07** | 6.38** |
| 种群内F值 F value within population | - | - | - | - | - | - | - |
| 性状 Traits | 方差分量Variance component | 方差分量百分比 Percentage of variance component | 表型分化系数 Phenotype of differentiation coefficient (%) | |||||
|---|---|---|---|---|---|---|---|---|
| 种群间 Among populations | 种群内 Within population | 机误 Errors | 种群间 Among populations | 种群内 Within population | 机误 Errors | |||
| SL | 0.001 882 | 0.009 550 | 0.031 017 | 4.434 2 | 22.497 1 | 73.068 6 | 16.462 | |
| SW | 0.000 372 | 0.002 477 | 0.006 498 | 3.978 0 | 26.504 5 | 69.517 5 | 13.057 | |
| SLW | 0.003 032 | 0.042 915 | 0.161 584 | 1.461 0 | 20.678 6 | 77.860 4 | 6.598 | |
| ST | 0.000 152 | 0.001 082 | 0.001 665 | 5.254 0 | 37.324 8 | 57.421 2 | 12.317 | |
| KT | 0.000 066 | 0.000 215 | 0.000 481 | 8.637 6 | 28.205 5 | 63.156 9 | 23.424 | |
| SKT | 0.001 130 | 0.032 541 | 0.418 792 | 0.249 8 | 7.192 0 | 92.558 3 | 3.356 | |
| 平均值Average | - | - | - | 3.442 3 | 20.345 1 | 76.212 7 | 11.885 | |
表3 印楝表型性状的方差分量及种群间表型分化系数
Table 3 Variance component of phenotypic traits among and within populations of Azadirachta indica
| 性状 Traits | 方差分量Variance component | 方差分量百分比 Percentage of variance component | 表型分化系数 Phenotype of differentiation coefficient (%) | |||||
|---|---|---|---|---|---|---|---|---|
| 种群间 Among populations | 种群内 Within population | 机误 Errors | 种群间 Among populations | 种群内 Within population | 机误 Errors | |||
| SL | 0.001 882 | 0.009 550 | 0.031 017 | 4.434 2 | 22.497 1 | 73.068 6 | 16.462 | |
| SW | 0.000 372 | 0.002 477 | 0.006 498 | 3.978 0 | 26.504 5 | 69.517 5 | 13.057 | |
| SLW | 0.003 032 | 0.042 915 | 0.161 584 | 1.461 0 | 20.678 6 | 77.860 4 | 6.598 | |
| ST | 0.000 152 | 0.001 082 | 0.001 665 | 5.254 0 | 37.324 8 | 57.421 2 | 12.317 | |
| KT | 0.000 066 | 0.000 215 | 0.000 481 | 8.637 6 | 28.205 5 | 63.156 9 | 23.424 | |
| SKT | 0.001 130 | 0.032 541 | 0.418 792 | 0.249 8 | 7.192 0 | 92.558 3 | 3.356 | |
| 平均值Average | - | - | - | 3.442 3 | 20.345 1 | 76.212 7 | 11.885 | |
| 性状 Trait | 种群间 Among populations | 种群内 Within population | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| WX | XBL | NMS | DHG | WW | NS | LD | MT | HCB | 平均值 Average | ||
| SL | 3.76 | 8.25 | 7.72 | 7.16 | 6.53 | 6.00 | 8.51 | 7.22 | 6.51 | 8.09 | 7.33hi |
| SW | 3.34 | 4.35 | 10.39 | 5.73 | 5.76 | 10.34 | 6.45 | 5.10 | 6.26 | 6.41 | 6.76i |
| SLW | 3.94 | 9.58 | 11.48 | 10.53 | 11.96 | 13.96 | 11.20 | 10.45 | 6.67 | 8.14 | 10.44fghi |
| ST | 7.98 | 19.43 | 19.65 | 13.40 | 19.71 | 14.24 | 16.32 | 11.70 | 13.14 | 18.59 | 16.24defg |
| KT | 9.04 | 14.40 | 15.05 | 14.64 | 11.46 | 10.05 | 17.33 | 11.10 | 17.98 | 15.90 | 14.21efg |
| SKT | 4.06 | 13.63 | 13.31 | 12.62 | 13.17 | 5.40 | 6.93 | 6.99 | 8.18 | 6.68 | 9.66ghi |
| SG | 19.62 | 19.63 | 22.39 | 15.54 | 12.84 | 21.23 | 17.08 | 14.55 | 10.34 | 19.91 | 17.06def |
| ZG | 52.49 | 85.98 | 85.85 | 95.78 | 96.53 | 47.63 | 36.35 | 51.52 | 46.23 | 85.63 | 70.17a |
| DZ | 13.86 | 26.57 | 17.09 | 10.93 | 24.65 | 31.10 | 30.82 | 20.16 | 27.29 | 22.74 | 23.48cd |
| GF | 22.59 | 33.38 | 26.84 | 22.05 | 21.09 | 28.62 | 25.35 | 25.05 | 30.53 | 18.90 | 25.76c |
| GS | 32.67 | 59.75 | 79.59 | 53.26 | 38.42 | 53.36 | 74.51 | 36.71 | 47.48 | 36.37 | 53.27b |
| azA | 8.94 | 22.28 | 17.74 | 13.92 | 8.86 | 7.66 | 13.68 | 16.33 | 9.47 | 13.34 | 13.70efgh |
| azB | 19.28 | 19.32 | 26.16 | 14.70 | 19.17 | 16.67 | 18.68 | 12.50 | 21.58 | 26.84 | 19.51cde |
| azAB | 10.30 | 19.99 | 19.47 | 13.33 | 9.13 | 6.58 | 12.52 | 12.70 | 7.18 | 12.82 | 12.64efghi |
| 平均值 Average | 15.13 | 25.47a | 26.62a | 21.68a | 21.38a | 19.49a | 21.12a | 17.29a | 18.49a | 21.45a | 21.44 |
表4 9个印楝实生种群的表型性状变异系数以及种群间和性状间Duncan检验变异系数的多重比较
Table 4 Variation coefficient of phenotypic traits in nine natural populations and Duncan’s test of CV among populations and phenotype trait of Azadirachta indica (%)
| 性状 Trait | 种群间 Among populations | 种群内 Within population | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| WX | XBL | NMS | DHG | WW | NS | LD | MT | HCB | 平均值 Average | ||
| SL | 3.76 | 8.25 | 7.72 | 7.16 | 6.53 | 6.00 | 8.51 | 7.22 | 6.51 | 8.09 | 7.33hi |
| SW | 3.34 | 4.35 | 10.39 | 5.73 | 5.76 | 10.34 | 6.45 | 5.10 | 6.26 | 6.41 | 6.76i |
| SLW | 3.94 | 9.58 | 11.48 | 10.53 | 11.96 | 13.96 | 11.20 | 10.45 | 6.67 | 8.14 | 10.44fghi |
| ST | 7.98 | 19.43 | 19.65 | 13.40 | 19.71 | 14.24 | 16.32 | 11.70 | 13.14 | 18.59 | 16.24defg |
| KT | 9.04 | 14.40 | 15.05 | 14.64 | 11.46 | 10.05 | 17.33 | 11.10 | 17.98 | 15.90 | 14.21efg |
| SKT | 4.06 | 13.63 | 13.31 | 12.62 | 13.17 | 5.40 | 6.93 | 6.99 | 8.18 | 6.68 | 9.66ghi |
| SG | 19.62 | 19.63 | 22.39 | 15.54 | 12.84 | 21.23 | 17.08 | 14.55 | 10.34 | 19.91 | 17.06def |
| ZG | 52.49 | 85.98 | 85.85 | 95.78 | 96.53 | 47.63 | 36.35 | 51.52 | 46.23 | 85.63 | 70.17a |
| DZ | 13.86 | 26.57 | 17.09 | 10.93 | 24.65 | 31.10 | 30.82 | 20.16 | 27.29 | 22.74 | 23.48cd |
| GF | 22.59 | 33.38 | 26.84 | 22.05 | 21.09 | 28.62 | 25.35 | 25.05 | 30.53 | 18.90 | 25.76c |
| GS | 32.67 | 59.75 | 79.59 | 53.26 | 38.42 | 53.36 | 74.51 | 36.71 | 47.48 | 36.37 | 53.27b |
| azA | 8.94 | 22.28 | 17.74 | 13.92 | 8.86 | 7.66 | 13.68 | 16.33 | 9.47 | 13.34 | 13.70efgh |
| azB | 19.28 | 19.32 | 26.16 | 14.70 | 19.17 | 16.67 | 18.68 | 12.50 | 21.58 | 26.84 | 19.51cde |
| azAB | 10.30 | 19.99 | 19.47 | 13.33 | 9.13 | 6.58 | 12.52 | 12.70 | 7.18 | 12.82 | 12.64efghi |
| 平均值 Average | 15.13 | 25.47a | 26.62a | 21.68a | 21.38a | 19.49a | 21.12a | 17.29a | 18.49a | 21.45a | 21.44 |
| 表型性状 Phenotypic trait | 主成分1 Principal component 1 | 主成分2 Principal component 2 | 主成分3 Principal component 3 | 主成分4 Principal component 4 | 主成分5 Principal component 5 |
|---|---|---|---|---|---|
| SL | 0.002 198 | -0.001 052 | 0.008 378 | 0.013 340 | 0.000 401 |
| SW | 0.000 218 | 0.000 513 | 0.009 554 | 0.002 810 | 0.006 186 |
| SLW | 0.001 647 | -0.002 110 | -0.015 717 | 0.008 465 | -0.007 330 |
| ST | 0.000 948 | 0.000 781 | 0.001 570 | 0.002 797 | -0.005 661 |
| KT | 0.000 324 | 0.000 033 | 0.002 127 | 0.002 771 | -0.003 080 |
| SKT | 0.001 725 | 0.005 915 | -0.007 598 | -0.014 930 | -0.022 047 |
| SG | 0.098 766 | 0.012 347 | 0.160 562 | -0.028 466 | 0.019 429 |
| ZG | 0.018 922 | 0.000 110 | 0.049 811 | -0.008 068 | -0.045 176 |
| DZ | 0.119 856 | 0.106 334 | 0.781 486 | 0.223 995 | -0.539 594 |
| GF | 0.079 642 | 0.059 780 | 0.438 917 | 0.338 906 | 0.816 078 |
| GS | 0.981 859 | -0.007 729 | -0.122 500 | -0.117 189 | 0.011 604 |
| azA | 0.034 673 | 0.444 442 | -0.276 853 | 0.541 106 | -0.035 571 |
| azB | -0.062 381 | 0.358 848 | 0.257 206 | -0.725 356 | 0.188 600 |
| azAB | -0.005 137 | 0.811 520 | -0.100 442 | -0.030 452 | -0.053 364 |
| 特征值 Characteristic root | 146.758 485 | 34.971 700 | 4.038 099 | 1.701 024 | 0.494 127 |
| 贡献率 Variance proportion (%) | 0.779 9 | 0.185 9 | 0.021 5 | 0.009 0 | 0.002 6 |
| 累计贡献率 Cumulative variance proportion (%) | 0.779 9 | 0.965 8 | 0.987 3 | 0.996 3 | 0.998 9 |
表5 前5个主成分的负荷量、特征值、贡献率和累积贡献率
Table 5 Factor loadings, characteristic root, variance proportion and cumulative variance proportion of five principal components
| 表型性状 Phenotypic trait | 主成分1 Principal component 1 | 主成分2 Principal component 2 | 主成分3 Principal component 3 | 主成分4 Principal component 4 | 主成分5 Principal component 5 |
|---|---|---|---|---|---|
| SL | 0.002 198 | -0.001 052 | 0.008 378 | 0.013 340 | 0.000 401 |
| SW | 0.000 218 | 0.000 513 | 0.009 554 | 0.002 810 | 0.006 186 |
| SLW | 0.001 647 | -0.002 110 | -0.015 717 | 0.008 465 | -0.007 330 |
| ST | 0.000 948 | 0.000 781 | 0.001 570 | 0.002 797 | -0.005 661 |
| KT | 0.000 324 | 0.000 033 | 0.002 127 | 0.002 771 | -0.003 080 |
| SKT | 0.001 725 | 0.005 915 | -0.007 598 | -0.014 930 | -0.022 047 |
| SG | 0.098 766 | 0.012 347 | 0.160 562 | -0.028 466 | 0.019 429 |
| ZG | 0.018 922 | 0.000 110 | 0.049 811 | -0.008 068 | -0.045 176 |
| DZ | 0.119 856 | 0.106 334 | 0.781 486 | 0.223 995 | -0.539 594 |
| GF | 0.079 642 | 0.059 780 | 0.438 917 | 0.338 906 | 0.816 078 |
| GS | 0.981 859 | -0.007 729 | -0.122 500 | -0.117 189 | 0.011 604 |
| azA | 0.034 673 | 0.444 442 | -0.276 853 | 0.541 106 | -0.035 571 |
| azB | -0.062 381 | 0.358 848 | 0.257 206 | -0.725 356 | 0.188 600 |
| azAB | -0.005 137 | 0.811 520 | -0.100 442 | -0.030 452 | -0.053 364 |
| 特征值 Characteristic root | 146.758 485 | 34.971 700 | 4.038 099 | 1.701 024 | 0.494 127 |
| 贡献率 Variance proportion (%) | 0.779 9 | 0.185 9 | 0.021 5 | 0.009 0 | 0.002 6 |
| 累计贡献率 Cumulative variance proportion (%) | 0.779 9 | 0.965 8 | 0.987 3 | 0.996 3 | 0.998 9 |
| SL | SW | SLW | ST | KT | SKT | SG | ZG | DZ | GF | GS | azA | azB | azAB | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SL | 1.000 00 | |||||||||||||
| SW | 0.357 73 | 1.000 00 | ||||||||||||
| SLW | 0.499 15 | -0.614 09* | 1.000 00 | |||||||||||
| ST | 0.801 89** | 0.191 87 | 0.463 28 | 1.000 00 | ||||||||||
| KT | 0.930 85** | 0.404 81 | 0.399 27 | 0.844 93** | 1.000 00 | |||||||||
| SKT | -0.499 75 | -0.232 70 | -0.235 14 | -0.211 65 | -0.591 62* | 1.000 00 | ||||||||
| SG | 0.709 79** | 0.277 27 | 0.286 88 | 0.774 39** | 0.558 43* | 0.113 27 | 1.000 00 | |||||||
| ZG | 0.913 34** | 0.247 49 | 0.524 23* | 0.861 73** | 0.877 99** | -0.369 59 | 0.795 22** | 1.000 00 | ||||||
| DZ | 0.545 71* | 0.623 44* | -0.143 28 | 0.680 70** | 0.541 38* | 0.157 23 | 0.771 40** | 0.555 84* | 1.000 00 | |||||
| GF | 0.575 45* | 0.646 69** | -0.098 31 | 0.558 06* | 0.452 25 | 0.015 26 | 0.736 05** | 0.481 05 | 0.833 87** | 1.000 00 | ||||
| GS | 0.504 81 | 0.088 19 | 0.250 68 | 0.660 08** | 0.333 53 | 0.246 57 | 0.927 45** | 0.594 45* | 0.619 39* | 0.611 84* | 1.000 00 | |||
| azA | -0.017 20 | 0.011 50 | 0.004 60 | 0.366 97 | 0.071 11 | 0.400 05 | 0.136 11 | 0.031 18 | 0.250 40 | 0.264 44 | 0.146 73 | 1.000 00 | ||
| azB | -0.330 11 | 0.184 31 | -0.349 87 | -0.037 89 | -0.156 17 | 0.320 34 | -0.173 48 | -0.132 21 | 0.126 97 | 0.035 27 | -0.304 65 | 0.612 22* | 1.000 00 | |
| azAB | -0.140 28 | 0.085 71 | -0.137 89 | 0.256 55 | 0.000 41 | 0.404 43 | 0.035 23 | -0.012 01 | 0.238 35 | 0.192 35 | -0.015 83 | 0.938 81** | 0.845 39** | 1.000 00 |
表6 印楝表型性状间的相关系数矩阵
Table 6 Coefficient matrix between phenotypic traits of Azadirachta indica
| SL | SW | SLW | ST | KT | SKT | SG | ZG | DZ | GF | GS | azA | azB | azAB | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SL | 1.000 00 | |||||||||||||
| SW | 0.357 73 | 1.000 00 | ||||||||||||
| SLW | 0.499 15 | -0.614 09* | 1.000 00 | |||||||||||
| ST | 0.801 89** | 0.191 87 | 0.463 28 | 1.000 00 | ||||||||||
| KT | 0.930 85** | 0.404 81 | 0.399 27 | 0.844 93** | 1.000 00 | |||||||||
| SKT | -0.499 75 | -0.232 70 | -0.235 14 | -0.211 65 | -0.591 62* | 1.000 00 | ||||||||
| SG | 0.709 79** | 0.277 27 | 0.286 88 | 0.774 39** | 0.558 43* | 0.113 27 | 1.000 00 | |||||||
| ZG | 0.913 34** | 0.247 49 | 0.524 23* | 0.861 73** | 0.877 99** | -0.369 59 | 0.795 22** | 1.000 00 | ||||||
| DZ | 0.545 71* | 0.623 44* | -0.143 28 | 0.680 70** | 0.541 38* | 0.157 23 | 0.771 40** | 0.555 84* | 1.000 00 | |||||
| GF | 0.575 45* | 0.646 69** | -0.098 31 | 0.558 06* | 0.452 25 | 0.015 26 | 0.736 05** | 0.481 05 | 0.833 87** | 1.000 00 | ||||
| GS | 0.504 81 | 0.088 19 | 0.250 68 | 0.660 08** | 0.333 53 | 0.246 57 | 0.927 45** | 0.594 45* | 0.619 39* | 0.611 84* | 1.000 00 | |||
| azA | -0.017 20 | 0.011 50 | 0.004 60 | 0.366 97 | 0.071 11 | 0.400 05 | 0.136 11 | 0.031 18 | 0.250 40 | 0.264 44 | 0.146 73 | 1.000 00 | ||
| azB | -0.330 11 | 0.184 31 | -0.349 87 | -0.037 89 | -0.156 17 | 0.320 34 | -0.173 48 | -0.132 21 | 0.126 97 | 0.035 27 | -0.304 65 | 0.612 22* | 1.000 00 | |
| azAB | -0.140 28 | 0.085 71 | -0.137 89 | 0.256 55 | 0.000 41 | 0.404 43 | 0.035 23 | -0.012 01 | 0.238 35 | 0.192 35 | -0.015 83 | 0.938 81** | 0.845 39** | 1.000 00 |
| 性状 Trait | 纬度 Latitude (N) | 经度 Longitude (E) | 海拔 Elevation (m) | 年平均气温 Annual mean air temperature (℃) | 年降水量 Annual precipitation (mm) |
|---|---|---|---|---|---|
| SL | -0.640 9* | 0.655 8* | -0.741 4* | 0.789 5** | 0.626 7 |
| SW | -0.335 2 | 0.260 6 | -0.283 3 | 0.237 9 | 0.271 6 |
| SLW | -0.187 3 | 0.238 6 | -0.302 5 | 0.371 8 | 0.201 9 |
| ST | -0.787 9** | 0.811 8** | -0.864 3** | 0.890 1** | 0.787 9** |
| KT | -0.615 2 | 0.682 3* | -0.713 1* | 0.774 1** | 0.675 6* |
| SKT | 0.244 6 | -0.317 3 | 0.269 5 | -0.300 7 | -0.354 0 |
| SG | -0.667 7* | 0.601 5 | -0.744 9* | 0.753 6* | 0.559 6 |
| ZG | -0.548 8 | 0.554 8 | -0.666 3* | 0.728 6* | 0.528 4 |
| DZ | -0.616 8 | 0.571 5 | -0.660 3* | 0.655 3* | 0.563 9 |
| GF | -0.747 9* | 0.583 8 | -0.700 0* | 0.612 0 | 0.550 1 |
| GS | -0.681 4* | 0.631 0 | -0.736 6* | 0.728 2* | 0.589 5 |
| azA | -0.339 3 | 0.234 0 | -0.219 9 | 0.119 1 | 0.174 8 |
| azB | 0.222 2 | -0.355 3 | 0.334 3 | -0.407 1 | -0.364 1 |
| azAB | -0.142 5 | 0.019 6 | -0.019 3 | -0.075 7 | -0.023 0 |
表7 印楝表型性状与地理因子的相关系数
Table 7 Coefficient between phenotypic traits and geographic factors of Azadirachta indica
| 性状 Trait | 纬度 Latitude (N) | 经度 Longitude (E) | 海拔 Elevation (m) | 年平均气温 Annual mean air temperature (℃) | 年降水量 Annual precipitation (mm) |
|---|---|---|---|---|---|
| SL | -0.640 9* | 0.655 8* | -0.741 4* | 0.789 5** | 0.626 7 |
| SW | -0.335 2 | 0.260 6 | -0.283 3 | 0.237 9 | 0.271 6 |
| SLW | -0.187 3 | 0.238 6 | -0.302 5 | 0.371 8 | 0.201 9 |
| ST | -0.787 9** | 0.811 8** | -0.864 3** | 0.890 1** | 0.787 9** |
| KT | -0.615 2 | 0.682 3* | -0.713 1* | 0.774 1** | 0.675 6* |
| SKT | 0.244 6 | -0.317 3 | 0.269 5 | -0.300 7 | -0.354 0 |
| SG | -0.667 7* | 0.601 5 | -0.744 9* | 0.753 6* | 0.559 6 |
| ZG | -0.548 8 | 0.554 8 | -0.666 3* | 0.728 6* | 0.528 4 |
| DZ | -0.616 8 | 0.571 5 | -0.660 3* | 0.655 3* | 0.563 9 |
| GF | -0.747 9* | 0.583 8 | -0.700 0* | 0.612 0 | 0.550 1 |
| GS | -0.681 4* | 0.631 0 | -0.736 6* | 0.728 2* | 0.589 5 |
| azA | -0.339 3 | 0.234 0 | -0.219 9 | 0.119 1 | 0.174 8 |
| azB | 0.222 2 | -0.355 3 | 0.334 3 | -0.407 1 | -0.364 1 |
| azAB | -0.142 5 | 0.019 6 | -0.019 3 | -0.075 7 | -0.023 0 |
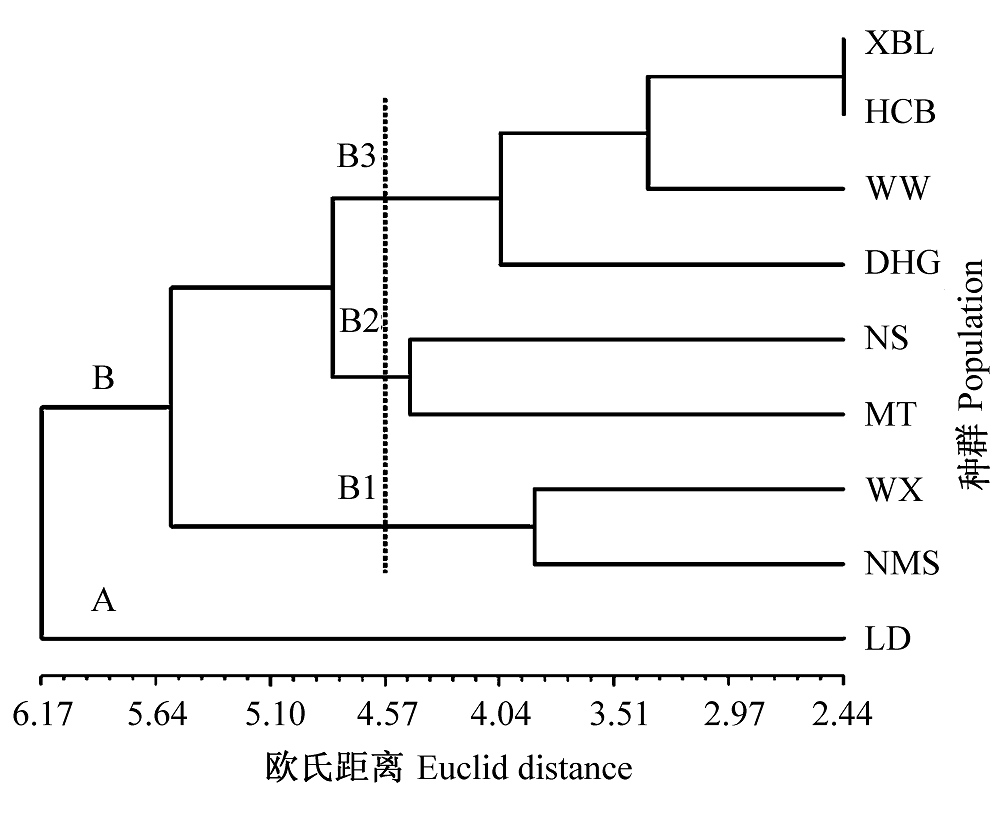
图1 印楝9个种群的表型性状聚类图。种群同表1。
Fig. 1 Un-weight pair-group method using arithmetic aver- ages (UPGMA) cluster based on phenotypic traits of nine populations of Azadirachta indica. Populations see Table 1.
| [1] | Chen YT ( 陈益泰), Li GY ( 李桂英), Wang HX ( 王惠雄) (1999). Study on phenotypic variation in natural range of long peduncled alder (Alnus cremastogyne). Forest Research (林业科学研究), 12, 379-385. (in Chinese with English abstract) |
| [2] | Cheng JH ( 程金焕) (2008). Variation and Influencing Factors of the Azadirachtin Content (印楝素含量变异及其影响因子). Master degree dissertation, Chinese Academy of Forestry, Beijing. 15-46. (in Chinese with English abstract) |
| [3] | Chinnamani S (1968). Cheap and effective method for establishing of farm woodlands in black cotton soils of Bellary. Indian Farming, 18, 3. |
| [4] | Chiu SF ( Zhao SH) ( 赵善欢), Zhang YG ( 张业光), Cai DZ ( 蔡德智), Lin GY ( 林冠亚) (1989). Preliminary report of neem tree introduction and cultivation in China. Journal of South China Agricultural University (华南农业大学学报), 10(2), 34-39. (in Chinese with English abstract) |
| [5] | Ellstrand NC, Elam DR (1993). Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics, 24, 217-242. |
| [6] | Feng SK ( 冯沙克), Zhao YF ( 赵元藩) (2007). Azadirachta indica’s growing in Yunnan and developmental status quo of relevant industries. Forest Inventory and Planning (林业调查规划), 32(5), 79-82. (in Chinese with English abstract) |
| [7] | Ge S ( 葛颂), Hong DY ( 洪德元) (1994). Genetic diversity and analysis (遗传多样性及其检测方法). In: Qian YQ (钱迎倩), Ma KP (马克平) eds. Theories and Applies for Biodiversity Research (生物多样性研究的理论与方法). Science and Technology Press, Beijing. 123-140. (in Chinese) |
| [8] | Ge S, Hong DY, Wang HQ, Liu ZY, Zhang CM (1998). Population genetic structure and conservation of an endangered conifer,Cathaya argyrophylla(Pinaceae) . International Journal of Plant Sciences, 159, 351-357. |
| [9] | Ge S ( 葛颂), Wang HQ ( 王海群), Zhang CM ( 张灿明), Hong DY ( 洪德元) (1997). Genetic diversity and population differentiation of Cathaya argyrophylla in Bamian Mountain. Acta Botanica Sinica (植物学报), 39, 266-271. (in Chinese with English abstract) |
| [10] | Ge S ( 葛颂), Wang MX ( 王明庥), Chen YW ( 陈岳武) (1988). An analysis of population genetic structure of masson pine by isozyme technique. Scientia Silvae Sinicae (林业科学), 24, 399-409. |
| [11] | Guo N ( 郭宁), Yang SH ( 杨树华), Ge WY ( 葛维亚), Ge H ( 葛红) (2011). Phenotypic diversity of natural populations of Rosa laxa Retz. in Tianshan Mountains of Xinjiang. Acta Horticulturae Sinica (园艺学报), 38, 495-502. (in Chinese with English abstract) |
| [12] | Hamrick JL, Godt MJW ( 1989). Isozymes and analyses of genetic structure of plant populations. In: Soltis DE, Soltis PS eds. Isozymes in Plant Biology. Dioscorides Press, Portland, Oregon, USA. 87-105. |
| [13] | Hamrick JL, Godt MJW (1990). Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL eds. Plant Population Genetics, Breeding, and Genetic Resources. Sinauer Association Inc., Massachusetts. 43-63. |
| [14] | Hamrick JL, Godt MJW, Sherman-Broyles SL (1992). Factors influencing levels of genetic diversity in woody plant species. New Forests, 6, 95-124. |
| [15] | He CZ ( 何承忠) (2005). Study on Genetic Diversity and Origin of Populus tomentosa (毛白杨遗传多样性及起源研究). PhD dissertation, Beijing Forestry University, Beijing. (in Chinese with English abstract) |
| [16] | Jin ZZ ( 金振洲), Ou XK ( 欧晓昆) (2000). Yuanjiang, Nujiang, Jinshajiang, Lancangjiang Vegetation of Dry-Hot Valley (元江、怒江、金沙江、澜沧江干热河谷植被). Yunnan University Press, Yunnan Science and Technology Press, Kunming. 291-297(in Chinese) |
| [17] | Kundu SK (1999a). Comparative analysis of seed morphometric and allozyme data among four populations of neem (Azadirachta indica). Genetic Resources and Crop Evolution, 46, 569-577. |
| [18] | Kundu SK (1999b). The mating system and genetic significance of polycarpy in the neem tree (Azadirachta indica). Theoretical and Applied Genetics, 99, 1216-1220. |
| [19] | Lai YQ ( 赖永祺) (2003). Cultivation of Neem (印楝栽培). Yunnan Science and Technology Press, Kunming. (in Chinese) |
| [20] |
Li B ( 李斌), Gu WC ( 顾万春) (2003). Review on genetic diversity in Pinus. Hereditas (遗传), 25, 740-748. (in Chinese with English abstract)
URL PMID |
| [21] | Li B ( 李斌), Gu WC ( 顾万春), Lu BM ( 卢宝明) (2002). A study on phenotypic diversity of seeds and cones characteristics in Pinus bungeana. Biodiversity Science (生物多样性), 10, 181-188. (in Chinese with English abstract) |
| [22] | Li CX ( 李长喜) (1998). A review of the studies on the phenotypic variation of forest trees in natural stands. Forest Research (林业科学研究), 11, 657-664. (in Chinese with English abstract) |
| [23] | Liu XL ( 刘新龙) (2004). Variation of Biological and Ecological Differences and AFLP Analysis Among Cultivated Neem Provenance (印楝(Azadirachta indica A. Juss)引种区不同种源的生物生态学差异及AFLP分析). Master degree dissertation, Yunnan University, Kunming. (in Chinese with English abstract) |
| [24] | Peng XM ( 彭兴民), Zhang YP ( 张燕平), Lai YQ ( 赖永祺), Zhao BR ( 赵保荣), Zhao PX ( 赵培仙) (2003). The biological characters of Azadirachta indica and result of introduction and cultural trail. Forest Research (林业科学研究), 16, 75-80. (in Chinese with English abstract) |
| [25] | Schaal BA, Leverich WJ, Rogstad SH (1991). Comparison of methods for assessing genetic variation in plant conservation biology. In: Falk DA, Holsinger KE eds. Genetics and Conservation of Rare Plants. Oxford University Press, New York. 123-134. |
| [26] | Schmutterer H, Ascher KRS, Isman MB (1995). The Neem Trees Azadirachta indica A. Juss. and Other Meliaceous Plants. VCH Vedagsgesellschaft, Weinheim, Germany. 1-29. |
| [27] | Venable DL (1984). Using intraspecific variation to study the ecological significance and evolution of plant life histories. In: Dirzo R, Sarukhan J eds. Perspectives on Plant Population Ecology. Sinauer, Sunderland. 166-187. |
| [28] | Wu JC ( 吴疆翀), Peng XM ( 彭兴民), Zheng YX ( 郑益兴), Zhang YP ( 张燕平) (2006). A preliminary study on the relationship between the Content of azadirachtin and the modality and the autumn of seed. Forest Research (林业科学研究), 19, 590-594. (in Chinese with English abstract) |
| [29] | Wu JC ( 吴疆翀), Peng XM ( 彭兴民), Zheng YX ( 郑益兴), Zhang YP ( 张燕平) (2008). Outcrossing rate and gene flow estimation of Azadirachta indica. Forest Research (林业科学研究), 21, 593-598. (in Chinese with English abstract) |
| [30] | Xu JR ( 续九如) (2006). Quantitative Genetics of Trees (林木数量遗传学). Higher Education Press, Beijing. (in Chinese) |
| [31] | Yang J ( 杨继) (1991a). Infraspecific variation in plant and the exploring methods. Journal of Wuhan Botanical Research (武汉植物学研究), 9, 185-195. (in Chinese with English abstract) |
| [32] | Yang J ( 杨继) (1991b). The applications of statistical methods to the study of intraspecific variation. Chinese Bulletin of Botany (植物学通报), 8, 30-36. (in Chinese with English abstract) |
| [33] | Zeng J ( 曾杰), Bai JY ( 白嘉雨) (2007). Some hot issues on phenotypic variation of natural plant populations. Guangxi Forestry Science (广西林业科学), 36, 65-70. (in Chinese with English abstract) |
| [34] |
Zeng J ( 曾杰), Zheng HS ( 郑海水), Gan SM ( 甘四明), Bai JY ( 白嘉雨) (2005). Phenotypic variation in natural populations of Betula alnoides in Guangxi, China. Scientia Silvae Sinicae (林业科学), 41(2), 59-65. (in Chinese with English abstract)
DOI URL |
| [35] | Zhang DQ ( 张德全), Yang YP ( 杨永平) (2008). A statistical and comparative analysis of genetic diversity detected by different molecular markers. Acta Botanica Yunnanica (云南植物研究), 30, 159-167. (in Chinese with English abstract) |
| [36] | Zhang YP ( 张燕平), Lai YQ ( 赖永祺), Peng XM ( 彭兴民), Liu J ( 刘娟) (2002). Global distribution and introduction cultivation state of Azadirachta indica. Forest Inventory and Planning (林业调查规划), 27(3), 98-101. (in Chinese with English abstract) |
| [37] | Zhao X ( 赵翾), Zhao SJ ( 赵树进) (2007). Phenotypic diversity of different Aquilaria sinensis (Lour.) Spreng. populations. Journal of South China University of Technology (Natural Science Edition) (华南理工大学学报 (自然科学版)), 35(4), 117-122, 133. (in Chinese with English abstract) |
| [38] | Zong QS ( 宗乾收), Lin J ( 林军), Wu YK ( 武永昆), Zeng W ( 曾文), Su PJ ( 苏鹏娟) (2003). Quantitative analysis of azadirachtin content of neem seeds by HPLC. Pesticides (农药), 42(4), 23-24. (in Chinese with English abstract) |
| [1] | 何庆海, 杨少宗, 李因刚, 沈鑫, 柳新红. 枫香树种群种子与果实表型性状变异分析[J]. 植物生态学报, 2018, 42(7): 752-763. |
| [2] | 柳江群, 尹明宇, 左丝雨, 杨绍斌, 乌云塔娜. 长柄扁桃天然种群表型变异[J]. 植物生态学报, 2017, 41(10): 1091-1102. |
| [3] | 尹明宇, 姜仲茂, 朱绪春, 包文泉, 赵罕, 乌云塔娜. 内蒙古山杏种群表型变异[J]. 植物生态学报, 2016, 40(10): 1090-1099. |
| [4] | 李因刚, 柳新红, 马俊伟, 石从广, 朱光权. 浙江楠种群表型变异[J]. 植物生态学报, 2014, 38(12): 1315-1324. |
| [5] | 陈天翌, 刘增辉, 娄安如. 刺萼龙葵种群在中国不同分布地区的表型变异[J]. 植物生态学报, 2013, 37(4): 344-353. |
| [6] | 李帅锋,苏建荣,刘万德,郎学东,张志钧,苏磊,贾呈鑫卓,杨华景. 思茅松天然群体种实表型变异[J]. 植物生态学报, 2013, 37(11): 998-1009. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
Copyright © 2022 版权所有 《植物生态学报》编辑部
地址: 北京香山南辛村20号, 邮编: 100093
Tel.: 010-62836134, 62836138; Fax: 010-82599431; E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
备案号: 京ICP备16067583号-19