

Chin J Plant Ecol ›› 2009, Vol. 33 ›› Issue (1): 1-11.DOI: 10.3773/j.issn.1005-264x.2009.01.001
Special Issue: 青藏高原植物生态学:种群生态学
• Research Articles • Next Articles
CHENG Kai1,2, SUN Kun3, WEN Hong-Yan4, ZHANG Min1, JIA Dong-Rui2, LIU Jian-Quan2,*( )
)
Received:2007-12-06
Accepted:2008-03-04
Online:2009-12-06
Published:2009-01-30
Contact:
LIU Jian-Quan
CHENG Kai, SUN Kun, WEN Hong-Yan, ZHANG Min, JIA Dong-Rui, LIU Jian-Quan. MATERNAL DIVERGENCE AND PHYLOGEOGRAPHICAL RELATIONSHIPS BETWEEN HIPPOPHAE GYANTSENSIS AND H. RHAMNOIDES SUBSP. YUNNANENSIS[J]. Chin J Plant Ecol, 2009, 33(1): 1-11.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.plant-ecology.com/EN/10.3773/j.issn.1005-264x.2009.01.001
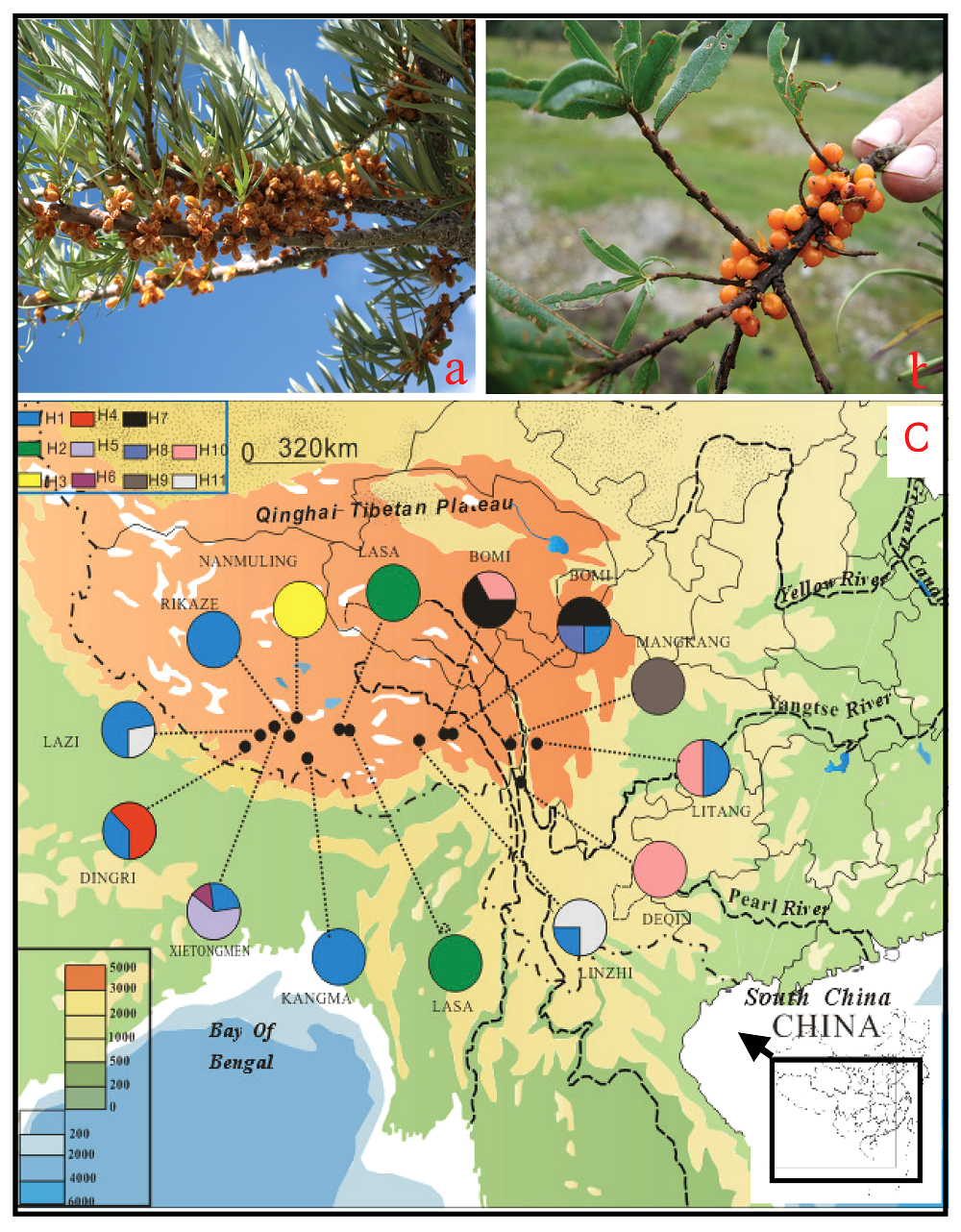
Fig. 1 Hippophae gyantsensis (from Nanmulin)(a), H. rhamnoides subsp. yunnanensis (from Bomi) (b) and the sampled populations and the recovered haplotypes in each population of two taxa (Lasa westward, H. gyantsensis and Linzhi eastward, H. rhamnoides subsp. yunnanensis)(c)
| 种群号 Pop. No. | 采集号 Pop. code | 采样地 Location | 纬度 Latitude | 经度 Longitude | 海拔 Alt. (m) | 样本数 N | 遗传多样性指数 h | 核苷酸多样性指数 π |
|---|---|---|---|---|---|---|---|---|
| 1 | Liu2006252 | 西藏定日 Dingri, Tibet | 28o40′ | 087o11′ | 4 370 | 8 | 0.536 | 0.004 65 |
| 2 | Liu2006261 | 西藏拉孜 Lazi, Tibet | 29o12′ | 087o41′ | 3 970 | 9 | 0.389 | 0.001 76 |
| 3 | Liu2006262 | 西藏谢通门 Xietongmen, Tibet | 29o25′ | 088o13′ | 3 940 | 8 | 0.429 | 0.005 75 |
| 4 | Liu2006263 | 西藏日喀则 Rikazi, Tibet | 29o21′ | 088o49′ | 3 860 | 8 | 0.000 | 0.000 00 |
| 5 | Liu2006217 | 西藏康马 Kangma, Tibet | 28o48′ | 089o40′ | 4 150 | 9 | 0.000 | 0.000 00 |
| 6 | Liu2006227 | 西藏南木林 Nanmulin, Tibet | 29o41′ | 089o10′ | 4 030 | 7 | 0.000 | 0.000 00 |
| 7 | Liu2006271 | 西藏拉萨 Lasa, Tibet | 29o40′ | 091o06′ | 3 650 | 6 | 0.000 | 0.000 00 |
| 8 | Liu2006274 | 西藏拉萨 Lasa, Tibet | 29o36′ | 091o08′ | 3 730 | 8 | 0.000 | 0.000 00 |
| 9 | Liu2006286 | 西藏林芝 Linzhi, Tibet | 29o44′ | 094o43′ | 3 640 | 8 | 0.429 | 0.001 94 |
| 10 | Liu2006211 | 西藏波密 Bomi, Tibet | 29o12′ | 095o54′ | 3 860 | 6 | 0.533 | 0.005 31 |
| 11 | Liu2006287 | 西藏波密 Bomi, Tibet | 29o45′ | 095o58′ | 2 950 | 8 | 0.714 | 0.005 86 |
| 12 | Liu2006304 | 西藏芒康 Mangkang, Tibet | 29o42′ | 098o24′ | 3 430 | 9 | 0.000 | 0.000 00 |
| 13 | Liu2006309 | 云南德钦 Deqin, Yunnan | 28o27′ | 098o53′ | 3 470 | 7 | 0.000 | 0.000 00 |
| 14 | Liu2006317 | 四川理塘 Litang, Sichuan | 29o43′ | 100o24′ | 3 640 | 8 | 0.571 | 0.010 36 |
Table 1 Origin of materials, estimates of haplotype diversity (h) and nucleotide diversity (π) in the 14 populations 1-8: Hippophae gyantsensis; 9-14: H. rhamnoides subsp. yunnanensis h: Haplotype diversity π: Nucleotide diversity
| 种群号 Pop. No. | 采集号 Pop. code | 采样地 Location | 纬度 Latitude | 经度 Longitude | 海拔 Alt. (m) | 样本数 N | 遗传多样性指数 h | 核苷酸多样性指数 π |
|---|---|---|---|---|---|---|---|---|
| 1 | Liu2006252 | 西藏定日 Dingri, Tibet | 28o40′ | 087o11′ | 4 370 | 8 | 0.536 | 0.004 65 |
| 2 | Liu2006261 | 西藏拉孜 Lazi, Tibet | 29o12′ | 087o41′ | 3 970 | 9 | 0.389 | 0.001 76 |
| 3 | Liu2006262 | 西藏谢通门 Xietongmen, Tibet | 29o25′ | 088o13′ | 3 940 | 8 | 0.429 | 0.005 75 |
| 4 | Liu2006263 | 西藏日喀则 Rikazi, Tibet | 29o21′ | 088o49′ | 3 860 | 8 | 0.000 | 0.000 00 |
| 5 | Liu2006217 | 西藏康马 Kangma, Tibet | 28o48′ | 089o40′ | 4 150 | 9 | 0.000 | 0.000 00 |
| 6 | Liu2006227 | 西藏南木林 Nanmulin, Tibet | 29o41′ | 089o10′ | 4 030 | 7 | 0.000 | 0.000 00 |
| 7 | Liu2006271 | 西藏拉萨 Lasa, Tibet | 29o40′ | 091o06′ | 3 650 | 6 | 0.000 | 0.000 00 |
| 8 | Liu2006274 | 西藏拉萨 Lasa, Tibet | 29o36′ | 091o08′ | 3 730 | 8 | 0.000 | 0.000 00 |
| 9 | Liu2006286 | 西藏林芝 Linzhi, Tibet | 29o44′ | 094o43′ | 3 640 | 8 | 0.429 | 0.001 94 |
| 10 | Liu2006211 | 西藏波密 Bomi, Tibet | 29o12′ | 095o54′ | 3 860 | 6 | 0.533 | 0.005 31 |
| 11 | Liu2006287 | 西藏波密 Bomi, Tibet | 29o45′ | 095o58′ | 2 950 | 8 | 0.714 | 0.005 86 |
| 12 | Liu2006304 | 西藏芒康 Mangkang, Tibet | 29o42′ | 098o24′ | 3 430 | 9 | 0.000 | 0.000 00 |
| 13 | Liu2006309 | 云南德钦 Deqin, Yunnan | 28o27′ | 098o53′ | 3 470 | 7 | 0.000 | 0.000 00 |
| 14 | Liu2006317 | 四川理塘 Litang, Sichuan | 29o43′ | 100o24′ | 3 640 | 8 | 0.571 | 0.010 36 |
| 核苷酸位点Site 单倍型 Haplotypes | 5 5 | 5 6 | 2 0 0 | 2 4 2 | 5 1 4 | 5 1 6 | 5 2 2 | 5 2 6 | 5 4 4 | 5 6 9 | 5 8 1 | 6 0 4 | 6 0 5 | 6 1 0 | 6 1 4 | 6 4 9 | 6 8 1 | 6 8 6 | 6 9 5 | 6 9 7 | 7 0 2 | 7 5 9 | 7 7 7 | 7 8 0 | 7 8 1 | 8 1 3 | 8 4 9 | 8 8 3 | 9 0 4 | 9 2 6 | 9 3 6 | 9 4 7 | 9 7 1 | 9 7 4 | 9 7 5 | 1 0 0 1 | 1 0 0 8 | 1 0 1 4 | 1 0 1 5 | 1 0 3 2 | 1 0 7 3 | 1110 | 1124 | 1219 | 1224 | 1237 | 1416 | 1435 | 1483 | 1 4 98 | 1 5 0 9 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | A | T | T | G | C | T | C | T | A | T | T | A | A | T | C | A | A | C | A | T | A | T | A | T | G | T | G | C | T | T | C | △ | ● | A | ■ | ★ | ◇ | A | ㊣ | # | ⊙ | A | G | C | T | - | T | C | A | A | C |
| H2 | A | T | T | A | C | T | C | T | A | T | C | A | A | T | C | A | A | G | A | T | A | T | A | T | G | T | G | C | T | T | C | △ | ● | C | ■ | ★ | ◇ | A | ㊣ | - | ⊙ | A | G | A | T | - | T | C | C | A | C |
| H3 | T | G | C | G | C | T | C | C | C | T | C | A | A | T | C | C | A | G | G | G | C | G | G | T | G | G | G | C | T | T | C | △ | ● | A | ■ | ★ | ◇ | A | ㊣ | # | ⊙ | A | G | C | T | - | T | C | A | A | C |
| H4 | T | G | C | G | C | T | C | C | C | T | C | A | A | A | A | A | C | C | A | G | A | T | G | G | G | T | G | C | T | T | C | △ | ● | A | ■ | ★ | ◇ | A | ㊣ | # | ⊙ | A | G | C | T | - | T | C | A | A | C |
| H5 | T | G | C | G | T | C | T | C | C | A | C | T | C | T | C | A | C | C | A | G | A | T | G | G | A | T | A | C | T | T | C | △ | ● | A | ■ | ★ | ◇ | A | ㊣ | # | ⊙ | A | G | C | T | - | T | C | A | A | C |
| H6 | T | G | C | G | T | C | T | C | C | A | C | T | C | T | C | A | C | C | A | G | A | T | G | G | A | T | A | C | T | T | C | △ | ● | A | ■ | ★ | ◇ | A | ㊣ | - | ⊙ | A | G | C | T | - | T | C | A | A | C |
| H7 | T | G | C | G | C | T | C | C | C | A | C | A | A | A | A | A | C | C | A | G | A | T | G | G | G | T | G | C | T | A | C | - | - | A | - | - | - | - | - | - | - | G | G | A | G | * | T | C | C | - | C |
| H8 | T | G | C | G | C | T | C | C | C | T | C | A | A | A | A | A | C | C | A | G | A | T | G | G | G | T | G | C | T | T | C | - | - | A | - | - | - | - | - | - | - | G | G | A | G | * | T | C | C | - | C |
| H9 | T | G | C | G | C | T | C | C | C | T | C | A | A | T | C | C | A | G | G | G | C | G | G | T | G | G | G | T | C | T | G | - | ● | A | ■ | - | ◇ | C | ㊣ | - | ⊙ | G | A | A | T | * | T | G | C | - | T |
| H10 | T | G | C | G | T | C | T | C | C | A | C | T | C | T | C | A | C | C | A | G | A | T | G | G | A | T | A | C | T | A | C | - | ● | C | ■ | ★ | ◇ | A | ㊣ | - | ⊙ | G | G | A | T | * | G | C | C | - | C |
| H11 | A | T | T | G | C | T | C | C | A | T | T | A | A | T | C | A | A | C | A | T | A | T | A | T | G | T | G | C | T | A | C | - | ● | C | ■ | ★ | ◇ | A | ㊣ | - | ⊙ | G | G | A | T | * | G | C | C | - | C |
Table 2 The sequence alignment of the 11 recovered haplotypes based trnL-F and trnS-G sequence in two taxa of Hippophae (△●■★◇㊣#⊙*, nine indels)
| 核苷酸位点Site 单倍型 Haplotypes | 5 5 | 5 6 | 2 0 0 | 2 4 2 | 5 1 4 | 5 1 6 | 5 2 2 | 5 2 6 | 5 4 4 | 5 6 9 | 5 8 1 | 6 0 4 | 6 0 5 | 6 1 0 | 6 1 4 | 6 4 9 | 6 8 1 | 6 8 6 | 6 9 5 | 6 9 7 | 7 0 2 | 7 5 9 | 7 7 7 | 7 8 0 | 7 8 1 | 8 1 3 | 8 4 9 | 8 8 3 | 9 0 4 | 9 2 6 | 9 3 6 | 9 4 7 | 9 7 1 | 9 7 4 | 9 7 5 | 1 0 0 1 | 1 0 0 8 | 1 0 1 4 | 1 0 1 5 | 1 0 3 2 | 1 0 7 3 | 1110 | 1124 | 1219 | 1224 | 1237 | 1416 | 1435 | 1483 | 1 4 98 | 1 5 0 9 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | A | T | T | G | C | T | C | T | A | T | T | A | A | T | C | A | A | C | A | T | A | T | A | T | G | T | G | C | T | T | C | △ | ● | A | ■ | ★ | ◇ | A | ㊣ | # | ⊙ | A | G | C | T | - | T | C | A | A | C |
| H2 | A | T | T | A | C | T | C | T | A | T | C | A | A | T | C | A | A | G | A | T | A | T | A | T | G | T | G | C | T | T | C | △ | ● | C | ■ | ★ | ◇ | A | ㊣ | - | ⊙ | A | G | A | T | - | T | C | C | A | C |
| H3 | T | G | C | G | C | T | C | C | C | T | C | A | A | T | C | C | A | G | G | G | C | G | G | T | G | G | G | C | T | T | C | △ | ● | A | ■ | ★ | ◇ | A | ㊣ | # | ⊙ | A | G | C | T | - | T | C | A | A | C |
| H4 | T | G | C | G | C | T | C | C | C | T | C | A | A | A | A | A | C | C | A | G | A | T | G | G | G | T | G | C | T | T | C | △ | ● | A | ■ | ★ | ◇ | A | ㊣ | # | ⊙ | A | G | C | T | - | T | C | A | A | C |
| H5 | T | G | C | G | T | C | T | C | C | A | C | T | C | T | C | A | C | C | A | G | A | T | G | G | A | T | A | C | T | T | C | △ | ● | A | ■ | ★ | ◇ | A | ㊣ | # | ⊙ | A | G | C | T | - | T | C | A | A | C |
| H6 | T | G | C | G | T | C | T | C | C | A | C | T | C | T | C | A | C | C | A | G | A | T | G | G | A | T | A | C | T | T | C | △ | ● | A | ■ | ★ | ◇ | A | ㊣ | - | ⊙ | A | G | C | T | - | T | C | A | A | C |
| H7 | T | G | C | G | C | T | C | C | C | A | C | A | A | A | A | A | C | C | A | G | A | T | G | G | G | T | G | C | T | A | C | - | - | A | - | - | - | - | - | - | - | G | G | A | G | * | T | C | C | - | C |
| H8 | T | G | C | G | C | T | C | C | C | T | C | A | A | A | A | A | C | C | A | G | A | T | G | G | G | T | G | C | T | T | C | - | - | A | - | - | - | - | - | - | - | G | G | A | G | * | T | C | C | - | C |
| H9 | T | G | C | G | C | T | C | C | C | T | C | A | A | T | C | C | A | G | G | G | C | G | G | T | G | G | G | T | C | T | G | - | ● | A | ■ | - | ◇ | C | ㊣ | - | ⊙ | G | A | A | T | * | T | G | C | - | T |
| H10 | T | G | C | G | T | C | T | C | C | A | C | T | C | T | C | A | C | C | A | G | A | T | G | G | A | T | A | C | T | A | C | - | ● | C | ■ | ★ | ◇ | A | ㊣ | - | ⊙ | G | G | A | T | * | G | C | C | - | C |
| H11 | A | T | T | G | C | T | C | C | A | T | T | A | A | T | C | A | A | C | A | T | A | T | A | T | G | T | G | C | T | A | C | - | ● | C | ■ | ★ | ◇ | A | ㊣ | - | ⊙ | G | G | A | T | * | G | C | C | - | C |
| 多样性参数 Diversity parameters | 合计 Total | 江孜沙棘 H. gyantsensis | 云南沙棘 H. rhamnoides subsp. yunnanensis |
|---|---|---|---|
| 种群 Populations | 15 | 8 | 6 |
| 单倍型 Haplotypes | 11 | 7 | 6 |
| HS | 0.270 (0.077 4) | 0.229(0.092 6) | 0.343 (0.147 1) |
| HT | 0.884 (0.043 8) | 0.850 (0.074 9) | 0.919 (0.065 5) |
| GST | 0.694 (0.090 2) | 0.730 (0.103 3) | 0.627 (0.169 9) |
| VS | 0.257 (0.079 6) | 0.166 (0.084 3) | 0.274 (0.131 6) |
| VT | 0.0934 (0.061 3) | 0.862 (0.126 0) | 1.036 (0.069 9) |
| NST | 0.725 (0.086 4) | 0.807 (0.108 0) | 0.735 (0.161 2) |
| NST -GST | 0.024 | 0.077 | 0.068 |
| h | 0.835 | 0.721 | 0.820 |
| π | 0.010 20 | 0.006 68 | 0.011 59 |
| Tajima’s D | 2.261 0* | 0..854 3 | 2.354 1* |
| Fu & Li’s D* | 2.083 8** | 1.883 7** | 1.908 6** |
Table 3 Genetic diversity and estimated indexes of two Hippophae taxa
| 多样性参数 Diversity parameters | 合计 Total | 江孜沙棘 H. gyantsensis | 云南沙棘 H. rhamnoides subsp. yunnanensis |
|---|---|---|---|
| 种群 Populations | 15 | 8 | 6 |
| 单倍型 Haplotypes | 11 | 7 | 6 |
| HS | 0.270 (0.077 4) | 0.229(0.092 6) | 0.343 (0.147 1) |
| HT | 0.884 (0.043 8) | 0.850 (0.074 9) | 0.919 (0.065 5) |
| GST | 0.694 (0.090 2) | 0.730 (0.103 3) | 0.627 (0.169 9) |
| VS | 0.257 (0.079 6) | 0.166 (0.084 3) | 0.274 (0.131 6) |
| VT | 0.0934 (0.061 3) | 0.862 (0.126 0) | 1.036 (0.069 9) |
| NST | 0.725 (0.086 4) | 0.807 (0.108 0) | 0.735 (0.161 2) |
| NST -GST | 0.024 | 0.077 | 0.068 |
| h | 0.835 | 0.721 | 0.820 |
| π | 0.010 20 | 0.006 68 | 0.011 59 |
| Tajima’s D | 2.261 0* | 0..854 3 | 2.354 1* |
| Fu & Li’s D* | 2.083 8** | 1.883 7** | 1.908 6** |
| 变异来源 Source of variation | 自由度 df | 方差 SS | 变异成分VC | 变异比例 Variation (%) | 固定指数 Fixation index | p |
|---|---|---|---|---|---|---|
| 类群间 Between taxa | 1 | 4.209 | 0.034 7 | 7.66 | FCT= 0.076 6 | 0.064 5 |
| 群体间 Among population within groups | 12 | 28.012 | 0.282 9 | 62.42 | FSC= 0.675 9 | < 0.001 |
| 群体内 Within populations | 95 | 12.889 | 0.135 7 | 29.92 | FST =0.700 8 | < 0.001 |
| 总计 Total | 108 | 45.110 | 0.453 4 |
Table 4 Analyses of molecular variance (AMOVAs) for all chlorotypes
| 变异来源 Source of variation | 自由度 df | 方差 SS | 变异成分VC | 变异比例 Variation (%) | 固定指数 Fixation index | p |
|---|---|---|---|---|---|---|
| 类群间 Between taxa | 1 | 4.209 | 0.034 7 | 7.66 | FCT= 0.076 6 | 0.064 5 |
| 群体间 Among population within groups | 12 | 28.012 | 0.282 9 | 62.42 | FSC= 0.675 9 | < 0.001 |
| 群体内 Within populations | 95 | 12.889 | 0.135 7 | 29.92 | FST =0.700 8 | < 0.001 |
| 总计 Total | 108 | 45.110 | 0.453 4 |
| 分支 Clades | Chi平方统计 Chi-square statistic | p | 分支检索 Clade key | 推论 Inferences |
|---|---|---|---|---|
| 1-1 | 16.000 0 | <0.001 | 1-19-20-2-11-17 NO | 趋势不明显 Inconclusive outcome. |
| 1-2 | 90.592 9 | <0.001 | 1-2-3-5-15NO | 片段化或长距离扩张 Past fragmentation and/or long distance colonization |
| 1-3 | 10.000 0 | 0.006 | 1-19-20-2-11-17 NO | 趋势不明显 Inconclusive outcome |
| 1-4 | 14.000 0 | 0.075 | 1-19-20-2-11-17 NO | 趋势不明显 Inconclusive outcome |
| 1-5 | 1.666 7 | 0.479 | 1-2-11-17 NO | 趋势不明显 Inconclusive outcome |
| 整个分枝 Total cladogram | 318.775 5 | <0.001 | 1-2-11-17 NO | 趋势不明显 Inconclusive outcome |
Table 6 The inference from the nested clade analysis following the given keys
| 分支 Clades | Chi平方统计 Chi-square statistic | p | 分支检索 Clade key | 推论 Inferences |
|---|---|---|---|---|
| 1-1 | 16.000 0 | <0.001 | 1-19-20-2-11-17 NO | 趋势不明显 Inconclusive outcome. |
| 1-2 | 90.592 9 | <0.001 | 1-2-3-5-15NO | 片段化或长距离扩张 Past fragmentation and/or long distance colonization |
| 1-3 | 10.000 0 | 0.006 | 1-19-20-2-11-17 NO | 趋势不明显 Inconclusive outcome |
| 1-4 | 14.000 0 | 0.075 | 1-19-20-2-11-17 NO | 趋势不明显 Inconclusive outcome |
| 1-5 | 1.666 7 | 0.479 | 1-2-11-17 NO | 趋势不明显 Inconclusive outcome |
| 整个分枝 Total cladogram | 318.775 5 | <0.001 | 1-2-11-17 NO | 趋势不明显 Inconclusive outcome |
| 零步 Zero-step | 一步 One-step | 两步 Two-step | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 单倍型 Haplotype | P | Dc | Dn | 分枝Clades | P | Dc | Dn | 分枝Clades | P | Dc | Dn | ||
| H11 | T | 4.308 S | 62.946S | 1-1 | T | 114.035 S | 114.462 S | 2-1 | T | 318.000 S | 333.484 S | ||
| H2 | T | 240.918 L | 310.847 L | ||||||||||
| H1 | I | 362.262 | 378.008 | 1-2 | I | 362.262 | 378.008 L | ||||||
| H9 | T | 0.000 S | 445.873 | 1-3 | T | 445.898 L | 447.296 L | ||||||
| H3 | T | 0.000 S | 445.923 L | ||||||||||
| I-T | - | 0.000 L | 0.050 L | ||||||||||
| I-T | 55.127 | 106.036 L | |||||||||||
| H4 | I | 0.000 S | 74.645 L | 1-4 | I | 63.913 S | 494.868 | 2-2 | T | 485.997 L | 480.024 L | ||
| H5 | I | 0.000 S | 55.864 S | ||||||||||
| H6 | I | 0.000 | 976.800 | 1-5 | T | 396.468 | 580.510 L | ||||||
| H10 | T | 91.000 | 127.444 | ||||||||||
| I-T | - | -91.000 | 849.356 | ||||||||||
| H7 | T | 0.000 | 108.520 | 1-6 | T | 341.659 | 343.437 S | ||||||
| H8 | I | 272.171 | 236.337 | ||||||||||
| I-T | - | 272.191 | 127.818 | ||||||||||
| I-T | -309.718 S | 13.138 | |||||||||||
Table 5 Nested clade analyses of the recovered haplotypes
| 零步 Zero-step | 一步 One-step | 两步 Two-step | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 单倍型 Haplotype | P | Dc | Dn | 分枝Clades | P | Dc | Dn | 分枝Clades | P | Dc | Dn | ||
| H11 | T | 4.308 S | 62.946S | 1-1 | T | 114.035 S | 114.462 S | 2-1 | T | 318.000 S | 333.484 S | ||
| H2 | T | 240.918 L | 310.847 L | ||||||||||
| H1 | I | 362.262 | 378.008 | 1-2 | I | 362.262 | 378.008 L | ||||||
| H9 | T | 0.000 S | 445.873 | 1-3 | T | 445.898 L | 447.296 L | ||||||
| H3 | T | 0.000 S | 445.923 L | ||||||||||
| I-T | - | 0.000 L | 0.050 L | ||||||||||
| I-T | 55.127 | 106.036 L | |||||||||||
| H4 | I | 0.000 S | 74.645 L | 1-4 | I | 63.913 S | 494.868 | 2-2 | T | 485.997 L | 480.024 L | ||
| H5 | I | 0.000 S | 55.864 S | ||||||||||
| H6 | I | 0.000 | 976.800 | 1-5 | T | 396.468 | 580.510 L | ||||||
| H10 | T | 91.000 | 127.444 | ||||||||||
| I-T | - | -91.000 | 849.356 | ||||||||||
| H7 | T | 0.000 | 108.520 | 1-6 | T | 341.659 | 343.437 S | ||||||
| H8 | I | 272.171 | 236.337 | ||||||||||
| I-T | - | 272.191 | 127.818 | ||||||||||
| I-T | -309.718 S | 13.138 | |||||||||||
| [1] | Avise JC (2000). Phylogeography: The History and Formation of Species. Harvard University Press, London. |
| [2] | Bartish IV, Jeppsson N, Bartish GI, Lu R, Nybom H (2000). Inter- and intraspecific genetic variation in Hippophae(Elaeagnaceae) investigated by RAPD markers. Plant Systematics and Evolution, 225, 81-101. |
| [3] | Bartish IV, Jeppsson N, Nybom H, Swenson U (2002). Phylogeny of Hippophae (Elaeagnaceae) inferred from parsimony analysis of chloroplast DNA and morphology. Systematic Botany, 27, 41-54. |
| [4] |
Bartish IV, Kadereit JW, Comes HP (2006). Late Quaternary history of Hippophae rhamnoides L. (Elaeagnaceae) inferred from chalcone synthase intron (Chsi) sequences and chloroplast DNA variation. Molecular Ecology, 15, 897-915.
DOI URL PMID |
| [5] |
Colosimo PF, Hosemann KE, Balabhadra S, Villarreal GJ, Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D, Kingsley DM (2005). Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science, 307, 1928-1933.
DOI URL PMID |
| [6] | Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochemical Bulletin, 19, 11-15. |
| [7] |
Excoffier L, Laval G, Schneider S (2005). ARLEQUIN (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics, 1, 47-50.
URL PMID |
| [8] |
Fu YX (1997). Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics, 147, 915-925.
URL PMID |
| [9] |
Hewitt G (2000). The genetic legacy of the Quaternary ice ages. Nature, 405, 907-913.
DOI URL PMID |
| [10] |
Hyvonen J (1996). On phylogeny of Hippophae(Elaeagnaceae). Nordic Journal of Botany, 16, 51-62.
DOI URL |
| [11] | Lian YS (廉永善) (2000). The Plant Biology and Chemistry of Hippophae (沙棘属植物生物学和化学). Gansu Science and Technology Press, Lanzhou. (in Chinese) |
| [12] | Lian YS, Chen XL, Lian H (1998). Systematic classification of the genus Hippophae L. Seabuckthorn Research, 1, 13-23. |
| [13] |
Meng LH (孟丽华), Yang HL (杨惠玲), Wu GL (吴桂莉), Wang YJ (王玉金) (2008). Phylogeography of Hippophae neurocarpa (Elaeagnaceae) inferred from the chloroplast DNA trnL-F sequence variation. Journal of Systematics and Evolution (植物分类学报), 46, 32-40. (in Chinese with English abstract)
DOI URL |
| [14] |
Newton AC, Allnot TR, Gillies ACM, Lowe AJ, Ennos RA (1999). Molecular phylogeography intraspecific variation and conservation of tree species. Trends in Ecology and Evolution, 14, 140-145.
DOI URL PMID |
| [15] |
Petit RJ, Aguinagalde I, de Beaulieu JL, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, Mohanty A, Muller-Starck GM, Demesure-Musch B, Palme A, Martin JP, Rendell S, Vendramin GG (2003). Glacial refugia: hotspots but not melting pots of genetic diversity. Science, 300, 1563-1565.
DOI URL PMID |
| [16] |
Pons O, Petit RJ (1995). Estimation, variance and optimal sampling of gene diversity. I. Haploid locus. Theoretical and Applied Genetics, 90, 462-470.
DOI URL PMID |
| [17] |
Posada D, Crandall KA, Templeton AR (2000). GEODIS: a program for the cladistic nested analysis of the geographical distribution of genetic haplotypes. Molecular Ecology, 9, 487-488.
DOI URL PMID |
| [18] |
Rieseberg LH (1997). Hybrid origins of plant species. Annual Review of Ecology and Systematics, 28, 359-389.
DOI URL |
| [19] |
Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, Schwarzbach AE, Donovan LA, Lexer C (2003). Major ecological transitions in annual sunflowers facilitated by hybridization. Science, 301, 1211-1216.
DOI URL PMID |
| [20] |
Rieseberg LH, Widmer A, Arntz MA, Burke JM (2002). Directional selection is the primary cause of phenotypic diversification. Proceedings of the National Academy of Sciences of the United States of America, 99, 12242-12245.
DOI URL PMID |
| [21] |
Rieseberg LH, Wood TE, Baack E (2006). The nature of plant species. Nature, 440, 524-527.
DOI URL PMID |
| [22] | Rousi A (1971). The genus Hippophae L. A taxonomic study. Annales Botanici Fennici, 8, 177-227. |
| [23] |
Rozas J, Sanchez-Delbarrio JC, Messeguer X, Rozas R (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics, 19, 2496-2497.
DOI URL PMID |
| [24] | Shi YF (施雅风), Li JJ (李吉均), Li BY (李炳元) (1998). Uplift and Environmental Changes of Qinghai-Tibetan Plateau in the Late Cenozoic (青藏高原晚新生代隆升与环境变化). Guangdong Science and Technology Press, Guangzhou. (in Chinse) |
| [25] |
Song BH, Wang XQ, Wang XR, Ding KY, Hong DY (2003). Cytoplasmic composition in Pinus densata and population establishment of the diploid hybrid pine. Molecular Ecology, 12, 2995-3001.
DOI URL PMID |
| [26] |
Sun K, Chen XL, Ma RJ, Li CB, Wang Q, Ge S (2002). Molecular phylogenetics of Hippophae L.(Elaeagnaceae) based on the internal transcribed spacer (ITS) sequences of nrDNA. Plant Systematics and Evolution, 235, 121-134.
DOI URL |
| [27] | Sun K, Ma RJ, Chen XL, Li CB, Ge S (2003). Hybrid origin of the diploid species Hippophae goniocarpa evidenced by the internal transcribed spacers (ITS) of nuclear rDNA. Belgian Journal of Botany, 136, 91-96. |
| [28] | Swofford DL (2000). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates. Sunderland, Massachusetts. |
| [29] |
Taberlet P, Gielly L, Pautou G, Bouvet J (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology, 17, 1105-1109.
DOI URL PMID |
| [30] |
Templeton AR, Crandall KA, Sing CF (1992). A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics, 132, 619-633.
URL PMID |
| [31] |
Thompson JD, Higgins DG, Gibson TJ (1994). CLUST- ALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673-4680.
DOI URL PMID |
| [32] |
Wang AL, Schluetz F, Liu JQ (2008). Molecular evidence for double maternal origins of the diploid hybrid Hippophae goniocarpa(Elaeagnaceae). Botanical Journal of the Linnean Society, 156, 111-118.
DOI URL |
| [33] |
Wang XR, Szmidt AE, Savolainen O (2001). Genetic composition and diploid hybrid speciation of a high mountain pine, Pinus densata, native to the Tibetan Plateau. Genetics, 159, 337-346.
URL PMID |
| [34] | Weir BS (1996). Genetic Data Analysis II. Sinauer & Associates, Sunderland, Massachusetts. |
| [35] |
Whibley AC, Langlade NB, Andalo C, Hanna AI, Bangham A, Thebaud C, Coen E (2006). Evolutionary paths underlying flower color variation in Antirrhinum. Science, 313, 963-967.
DOI URL PMID |
| [36] |
Zhang Q (张茜), Yang R (杨锐), Wang Q (王钦), Liu JQ (刘建全) (2005). Phylogeography of Juniperus przewalskii (Cupressaceae) inferred from the chloroplast DNA trnT-trnF sequence variation. Acta Phytotaxonomica Sinica (植物分类学报), 43, 503-512. (in Chinese with English abstract)
DOI URL |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Chinese Journal of Plant Ecology
Tel: 010-62836134, 62836138, E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn