

植物生态学报 ›› 2012, Vol. 36 ›› Issue (8): 880-890.DOI: 10.3724/SP.J.1258.2012.00880
所属专题: 生物多样性
邹媛媛1, 刘琳1, 刘洋1, 赵亮1, 邓启云2,*( ), 吴俊2, 庄文2, 宋未1,*(
), 吴俊2, 庄文2, 宋未1,*( )
)
收稿日期:2011-12-09
接受日期:2012-04-25
出版日期:2012-12-09
发布日期:2012-08-21
通讯作者:
邓启云,宋未
作者简介:* E-mail: songwei@mail.cnu.edu.cn;
ZOU Yuan-Yuan1, LIU Lin1, LIU Yang1, ZHAO Liang1, DENG Qi-Yun2,*( ), WU Jun2, ZHUANG Wen2, SONG Wei1,*(
), WU Jun2, ZHUANG Wen2, SONG Wei1,*( )
)
Received:2011-12-09
Accepted:2012-04-25
Online:2012-12-09
Published:2012-08-21
Contact:
DENG Qi-Yun,SONG Wei
摘要:
种子固有细菌是植物内生细菌的重要来源, 对植物的健康以及接种细菌的定殖能产生重要影响。该文以杂交水稻(Oryza sativa)种子为研究对象, 比较研究了不同品种水稻种子中固有细菌群落的多样性。利用799f和1492r这对引物成功地从水稻种子中扩增出固有细菌16S rDNA片段; 通过构建16S rDNA文库和扩增核糖体RNA基因酶切分型(ARDRA)的方法, 对杂交水稻 ‘丰优611’ (‘丰源A’ × ‘远恢611’)、‘金优611’ (‘金23A’ × ‘远恢611’)和‘金23A/09H013’ ( ‘金23A’ × ‘09H013’) 3个组合的子代及其各自亲本的种子固有细菌群落结构的多样性进行了研究。构建的7个克隆文库中, 每个文库含有200-300个克隆, 30-40个操作分类单元(OTU), 对ARDRA分型得到的代表序列进行分析, 在16S rDNA文库中发现多种细菌类群, 包括α变形杆菌(α-Proteobacteria)、β变形杆菌、γ变形杆菌、放线菌(Actinobacteria)、厚壁菌(Firmicutes)和拟菌(Bacteroidetes), 优势菌属是泛菌属(Pantoea)和芽孢杆菌属(Bacillus)。不同品种的水稻种子固有细菌群落结构不同, 而杂交子代种子中的优势菌与亲本种子中的优势菌在种类和数量上都具有一定的相关性。此外, 子代种子中丰度5%以上的细菌也能在各自父本或母本中检测到。
邹媛媛, 刘琳, 刘洋, 赵亮, 邓启云, 吴俊, 庄文, 宋未. 不同水稻品种种子固有细菌群落的多样性. 植物生态学报, 2012, 36(8): 880-890. DOI: 10.3724/SP.J.1258.2012.00880
ZOU Yuan-Yuan, LIU Lin, LIU Yang, ZHAO Liang, DENG Qi-Yun, WU Jun, ZHUANG Wen, SONG Wei. Diversity of indigenous bacterial communities in Oryza sativa seeds of different varieties. Chinese Journal of Plant Ecology, 2012, 36(8): 880-890. DOI: 10.3724/SP.J.1258.2012.00880
| 杂交子代 Hybrid lines | 母本 Female parental lines | 父本 Male parental lines |
|---|---|---|
| ‘丰优611’ ‘Fengyou 611’ | ‘丰源A’ ‘Fengyuan A’ | ‘远恢611’ ‘Yuanhui 611’ |
| ‘金优611’ ‘Jinyou 611’ | ‘金23A’ ‘Jin 23A’ | ‘远恢611’ ‘Yuanhui 611’ |
| ‘金23A/09H013’ ‘Jin 23A/09H013’ | ‘金23A’ ‘Jin 23A’ | ‘09H013’ ‘09H013’ |
表1 3个杂交组合中7个水稻品种之间的遗传关系
Table 1 Genetic relationship among seven rice varieties of three hybrid combinations
| 杂交子代 Hybrid lines | 母本 Female parental lines | 父本 Male parental lines |
|---|---|---|
| ‘丰优611’ ‘Fengyou 611’ | ‘丰源A’ ‘Fengyuan A’ | ‘远恢611’ ‘Yuanhui 611’ |
| ‘金优611’ ‘Jinyou 611’ | ‘金23A’ ‘Jin 23A’ | ‘远恢611’ ‘Yuanhui 611’ |
| ‘金23A/09H013’ ‘Jin 23A/09H013’ | ‘金23A’ ‘Jin 23A’ | ‘09H013’ ‘09H013’ |
| ‘丰优611’ ‘Fengyou 611’ | ‘金优611’ ‘Jinyou 611’ | ‘金23A/09H013’ ‘Jin 23A/09H013’ | ‘丰源A’ ‘Fengyuan A’ | ‘金23A’ ‘Jin 23A’ | ‘远恢611’ ‘Yuanhui 611’ | ‘09H013’ | |
|---|---|---|---|---|---|---|---|
| 克隆数 No. of clones | 201 | 268 | 253 | 241 | 278 | 240 | 273 |
| OTU数 No. of OTUs | 30 | 38 | 39 | 42 | 43 | 41 | 38 |
| C值 C value | 93.03% | 93.66% | 93.28% | 95.02% | 93.88% | 92.08% | 92.31% |
表2 7个不同水稻品种的种子固有细菌16S rDNA文库评价
Table 2 Estimation of seeds’ indigenous bacterial 16S rDNA libraries of seven different rice varieties
| ‘丰优611’ ‘Fengyou 611’ | ‘金优611’ ‘Jinyou 611’ | ‘金23A/09H013’ ‘Jin 23A/09H013’ | ‘丰源A’ ‘Fengyuan A’ | ‘金23A’ ‘Jin 23A’ | ‘远恢611’ ‘Yuanhui 611’ | ‘09H013’ | |
|---|---|---|---|---|---|---|---|
| 克隆数 No. of clones | 201 | 268 | 253 | 241 | 278 | 240 | 273 |
| OTU数 No. of OTUs | 30 | 38 | 39 | 42 | 43 | 41 | 38 |
| C值 C value | 93.03% | 93.66% | 93.28% | 95.02% | 93.88% | 92.08% | 92.31% |
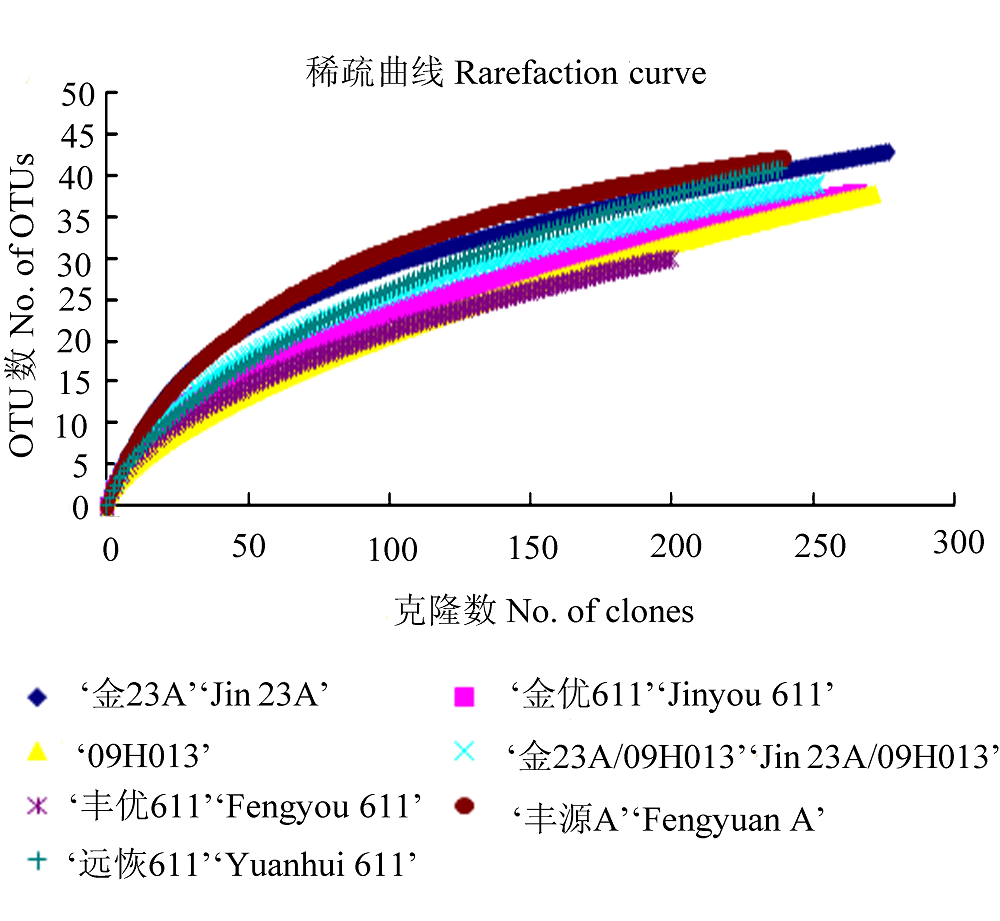
图1 7个不同水稻品种的种子固有细菌16S rDNA克隆文库的稀疏曲线。OTU, 操作分类单元。
Fig. 1 Rarefaction curves of seeds’ indigenous bacterial 16S rDNA clone libraries of seven different rice varieties. OTU, operational taxonomic unit.
| 类群 Group | 菌属 Genus | ‘丰优611’ ‘Fengyou 611’ | ‘金优611’ ‘Jinyou 611’ | ‘金23A/09H013’ ‘Jin 23A/09H013’ | ‘丰源A’ ‘FengyuanA’ | ‘金23A’ ‘Jin 23A’ | ‘远恢611’ ‘Yuanhui 611’ | ‘09H013’ |
|---|---|---|---|---|---|---|---|---|
| α变形杆菌 α-Proteobacteria | 17.41 | 3.36 | 28.06 | 16.18 | 1.44 | 35.42 | 27.84 | |
| 甲基杆菌 Methylobacterium | 6.47 | 0.75 | 9.49 | 0.41 | - | 2.92 | 0.73 | |
| 鞘氨醇单胞菌 Sphingomonas | 6.47 | 0.37 | 4.35 | 0.83 | 1.44 | 5.83 | 2.93 | |
| 土壤杆菌 Agrobacterium | 4.48 | 1.49 | 13.44 | 10.37 | - | 12.50 | 9.89 | |
| 橙单胞菌 Aurantimonas | - | - | - | - | - | - | 14.29 | |
| 沃尔巴克氏体 Wolbachia | - | - | - | - | - | 13.33 | - | |
| 其他 Others | - | 0.75 | 0.79 | 4.56 | - | 0.83 | - | |
| β变形杆菌 β-Proteobacteria | 2.49 | - | 2.77 | 22.41 | 0.72 | 0.83 | 7.69 | |
| 嗜甲基菌 Methylophilus | - | - | - | 8.30 | - | - | - | |
| 伯克霍尔德氏菌 Burkholderia | - | - | - | 5.81 | - | - | - | |
| 其他 Others | 2.49 | - | 2.77 | 8.30 | 0.72 | 0.83 | 7.69 | |
| γ变形杆菌 γ-Proteobacteria | 78.61 | 35.82 | 40.71 | 34.44 | 61.51 | 45.83 | 56.78 | |
| 泛菌 Pantoea | 64.18 | 5.60 | 31.62 | 22.82 | 7.55 | 40.00 | 54.95 | |
| 假单胞菌 Pseudomonas | 13.43 | 27.99 | 5.93 | 2.07 | 13.31 | 3.75 | - | |
| 肠杆菌 Enterobacter | - | - | - | - | 17.63 | - | - | |
| 欧文氏菌 Erwinia | - | 1.12 | - | - | 9.35 | - | - | |
| 不动杆菌 Acinetobacter | - | 0.37 | - | - | 7.20 | - | - | |
| 其他 Others | 1.00 | 0.74 | 3.16 | 9.54 | 6.47 | 2.08 | 1.83 | |
| 放线菌 Actinobacteria | 1.00 | 1.49 | 24.90 | 15.77 | 5.76 | 8.75 | 4.03 | |
| 微杆菌 Microbacterium | - | 1.12 | 19.37 | 2.07 | - | 7.50 | 4.03 | |
| 短小杆菌 Curtobacterium | 1.00 | 0.37 | 5.53 | 10.79 | 5.76 | 1.25 | - | |
| 其他 Others | - | - | - | 2.90 | - | - | - | |
| 厚壁菌 Firmicutes | - | 57.84 | 0.40 | 3.32 | 29.50 | - | 0.37 | |
| 芽孢杆菌 Bacillus | - | 57.84 | - | - | 28.78 | - | - | |
| 其他 Others | - | - | 0.40 | 3.32 | 0.72 | - | 0.37 | |
| 拟菌 Bacteroidetes | 0.50 | - | 3.16 | 4.98 | - | 9.17 | 3.30 | |
| 鞘胺醇杆菌 Sphingobacterium | - | - | - | - | - | 7.08 | 1.47 | |
| 其他 Others | 0.50 | - | 3.16 | 4.98 | - | 2.08 | 1.83 | |
| 其他 Others | - | 1.49 | - | 2.90 | 1.08 | - | - |
表3 三个杂交水稻品种及其亲本的种子固有细菌16S rDNA克隆文库中检测到的主要菌属的丰度
Table 3 Abundance of main genera detected in the seeds’ indigenous bacterial 16S rDNA clone libraries of three hybrid rice varieties and their parents (%)
| 类群 Group | 菌属 Genus | ‘丰优611’ ‘Fengyou 611’ | ‘金优611’ ‘Jinyou 611’ | ‘金23A/09H013’ ‘Jin 23A/09H013’ | ‘丰源A’ ‘FengyuanA’ | ‘金23A’ ‘Jin 23A’ | ‘远恢611’ ‘Yuanhui 611’ | ‘09H013’ |
|---|---|---|---|---|---|---|---|---|
| α变形杆菌 α-Proteobacteria | 17.41 | 3.36 | 28.06 | 16.18 | 1.44 | 35.42 | 27.84 | |
| 甲基杆菌 Methylobacterium | 6.47 | 0.75 | 9.49 | 0.41 | - | 2.92 | 0.73 | |
| 鞘氨醇单胞菌 Sphingomonas | 6.47 | 0.37 | 4.35 | 0.83 | 1.44 | 5.83 | 2.93 | |
| 土壤杆菌 Agrobacterium | 4.48 | 1.49 | 13.44 | 10.37 | - | 12.50 | 9.89 | |
| 橙单胞菌 Aurantimonas | - | - | - | - | - | - | 14.29 | |
| 沃尔巴克氏体 Wolbachia | - | - | - | - | - | 13.33 | - | |
| 其他 Others | - | 0.75 | 0.79 | 4.56 | - | 0.83 | - | |
| β变形杆菌 β-Proteobacteria | 2.49 | - | 2.77 | 22.41 | 0.72 | 0.83 | 7.69 | |
| 嗜甲基菌 Methylophilus | - | - | - | 8.30 | - | - | - | |
| 伯克霍尔德氏菌 Burkholderia | - | - | - | 5.81 | - | - | - | |
| 其他 Others | 2.49 | - | 2.77 | 8.30 | 0.72 | 0.83 | 7.69 | |
| γ变形杆菌 γ-Proteobacteria | 78.61 | 35.82 | 40.71 | 34.44 | 61.51 | 45.83 | 56.78 | |
| 泛菌 Pantoea | 64.18 | 5.60 | 31.62 | 22.82 | 7.55 | 40.00 | 54.95 | |
| 假单胞菌 Pseudomonas | 13.43 | 27.99 | 5.93 | 2.07 | 13.31 | 3.75 | - | |
| 肠杆菌 Enterobacter | - | - | - | - | 17.63 | - | - | |
| 欧文氏菌 Erwinia | - | 1.12 | - | - | 9.35 | - | - | |
| 不动杆菌 Acinetobacter | - | 0.37 | - | - | 7.20 | - | - | |
| 其他 Others | 1.00 | 0.74 | 3.16 | 9.54 | 6.47 | 2.08 | 1.83 | |
| 放线菌 Actinobacteria | 1.00 | 1.49 | 24.90 | 15.77 | 5.76 | 8.75 | 4.03 | |
| 微杆菌 Microbacterium | - | 1.12 | 19.37 | 2.07 | - | 7.50 | 4.03 | |
| 短小杆菌 Curtobacterium | 1.00 | 0.37 | 5.53 | 10.79 | 5.76 | 1.25 | - | |
| 其他 Others | - | - | - | 2.90 | - | - | - | |
| 厚壁菌 Firmicutes | - | 57.84 | 0.40 | 3.32 | 29.50 | - | 0.37 | |
| 芽孢杆菌 Bacillus | - | 57.84 | - | - | 28.78 | - | - | |
| 其他 Others | - | - | 0.40 | 3.32 | 0.72 | - | 0.37 | |
| 拟菌 Bacteroidetes | 0.50 | - | 3.16 | 4.98 | - | 9.17 | 3.30 | |
| 鞘胺醇杆菌 Sphingobacterium | - | - | - | - | - | 7.08 | 1.47 | |
| 其他 Others | 0.50 | - | 3.16 | 4.98 | - | 2.08 | 1.83 | |
| 其他 Others | - | 1.49 | - | 2.90 | 1.08 | - | - |
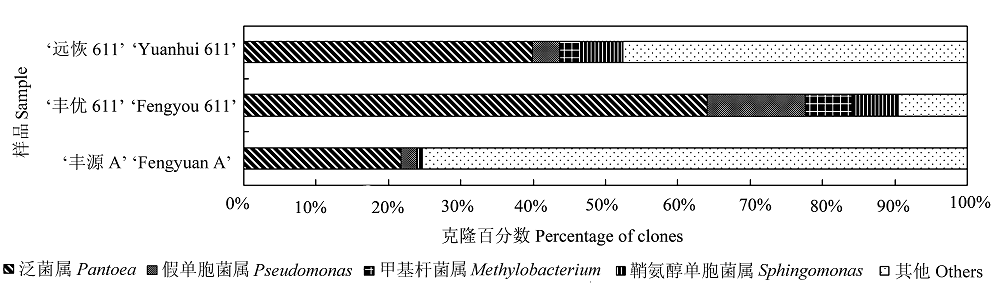
图2 ‘丰优611’及其亲本‘远恢611’和‘丰源A’种子中主要固有细菌类群的比较。
Fig. 2 Comparison of main indigenous bacterial groups in ‘Fengyou 611’ seed to its parental lines ‘Yuanhui 611’ and ‘Fengyuan A’.
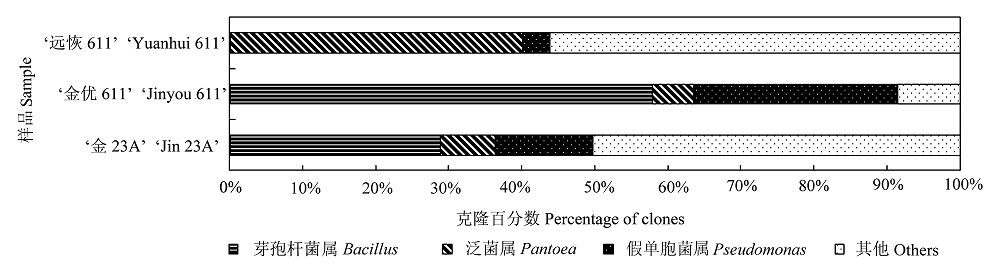
图3 ‘金优611’及其亲本‘远恢611’和‘金23A’种子中主要固有细菌类群的比较。
Fig. 3 Comparison of main indigenous bacterial groups in ‘Jinyou 611’ seed to its parental lines ‘Yuanhui 611’ and ‘Jin 23A’.
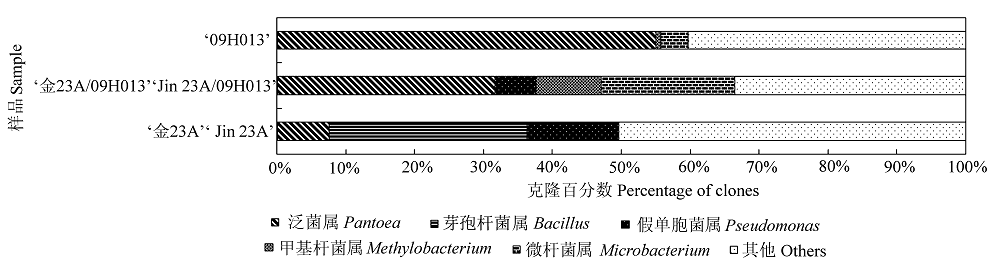
图4 ‘金23A/09H013’及其亲本‘09H013’和‘金23A’种子中主要固有细菌类群的比较。
Fig. 4 Comparison of main indigenous bacterial groups in ‘Jin 23A/09H013’ seed to its parental lines ‘09H013’ and ‘Jin 23A’.
| [1] | Adams PD, Kloepper JW (2002). Effect of host genotype on indigenous bacterial endophytes of cotton (Gossypium hirsutum L.). Plant and Soil, 240, 181-189. |
| [2] | Aulakh MS, Wassmann R, Bueno C, Kreuzwieser J, Rennenberg H (2001). Characterization of root exudates at different growth stages of ten rice (Oryza sativa L.) cultivars. Plant Biology, 3, 139-148. |
| [3] | Bacilio-Jiménez M, Aguilar-Flores S, del Valle MV, Pérez A, Zepeda A, Zenteno E (2001). Endophytic bacteria in rice seeds inhibit early colonization of roots by Azospirillum brasilense. Soil Biology & Biochemistry, 33, 167-172. |
| [4] |
Bayliss C, Bent E, Culham DE, MacLellan S, Clarke AJ, Wood JM, Brown GL (1997). Bacterial genetic loci implicated in the Pseudomonas putida GR12-2R3―canola mutualism: identification of an exudate-inducible sugar transporter. Canadian Journal of Microbiology, 43, 809-818.
DOI URL |
| [5] | Bouzar H, Chilton WS, Nesme X, Dessaux Y, Vaudequin V, Petit A, Jones JB, Hodge NC (1995). A new Agrobac- terium strain isolated from aerial tumors on Ficus benjamina L. Applied and Environmental Microbiology, 61, 65-73. |
| [6] | Bull CT, Weller DM, Thomashow LS (1991). Relationship between root colonization and suppression of Gaeumann- omyces graminis var. tritici by Pseudomonas fluorescens Strain 2-79. Phytopathology, 81, 954-959. |
| [7] | Buyer JS, Roberts DP, Russek-Cohen E (1999). Microbial community structure and function in the spermosphere as affected by soil and seed type. Canadian Journal of Microbiology, 45, 138-144. |
| [8] | Cankar K, Kraigher H, Ravnikar M, Rupnik M (2005). Bacterial endophytes from seeds of Norway spruce (Picea abies (L.) Karst.). FEMS Microbiology Letters, 244, 341-345. |
| [9] | Chakraborty U, Chakraborty B, Basnet M (2006). Plant growth promotion and induction of resistance in Camellia sinensis by Bacillus megaterium. Journal of Basic Microbiology, 46, 186-195. |
| [10] | Cruz AT, Cazacu AC, Allen CH (2007). Pantoea agglomerans, a plant pathogen causing human disease. Journal of Clinical Microbiology, 45, 1989-1992. |
| [11] | Dunleavy JM (1989). Curtobacterium plantarum sp. nov. is ubiquitous in plant leaves and is seed transmitted in soybean and corn. International Journal of Systematic and Evolutionary Microbiology, 39, 240-249. |
| [12] | Dussart-Baptista L, Bodilis J, Barray S, Frébourg N, Fournier M, Dupont JP, Jouenne T (2007). Recurrent recovery of Pseudomonas oryzihabitans strains in a karstified chalk aquifer. Water Research, 41, 111-117. |
| [13] | Espinosa-Urgel M, Ramos JL (2001). Expression of a Pseudomonas putida aminotransferase involved in lysine catabolism is induced in the rhizosphere. Applied and Environmental Microbiology, 67, 5219-5224. |
| [14] | Feng YJ, Shen DL, Dong XZ, Song W (2003). In vitro symplasmata formation in the rice diazotrophic endophyte Pantoea agglomerans YS19. Plant and Soil, 255, 435-444. |
| [15] | Ferreira A, Quecine MC, Lacava PT, Oda S, Azevedo JL, Araújo WL (2008). Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiology Letters, 287, 8-14. |
| [16] | Gitaitis R, Walcott R (2007). The epidemiology and manag- ement of seedborne bacterial diseases. Annual Review of Phytopathology, 45, 371-397. |
| [17] | Good IJ (1953). The population frequencies of species and the estimation of population parameters. Biometrika, 40, 237-264. |
| [18] | Grum M, Camloh M, Rudolph K, Ravnikar M (1998). Elimination of bean seed-borne bacteria by thermotherapy and meristem culture . Plant Cell, Tissue and Organ Culture, 52, 79-82. |
| [19] | Haas D, Défago G (2005). Biological control of soil-borne pathogens by Fluorescent pseudomonods. Nature Reviews Microbiology, 3, 307-319. |
| [20] |
Hallmann J, QuadtHallmann A, Mahaffee WF, Kloepper JW (1997). Bacterial endophytes in agricultural crops. Canadian Journal of Microbiology, 43, 895-914.
DOI URL |
| [21] | Hardoim PR, van Overbeek LS, van Elsas JD (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends in Microbiology, 16, 463-471. |
| [22] | Kaku H (2004). Histopathology of red stripe of rice. Plant Disease, 88, 1304-1309. |
| [23] | Kratz A, Greenberg D, Barki Y, Cohen E, Lifshitz M (2003). Pantoea agglomerans as a cause of septic arthritis after palm tree thorn injury; case report and literature review. Archives of Disease in Childhood, 88, 542-544. |
| [24] | Lidstrom ME, Chistoserdova L (2002). Plants in the pink: cytokinin production by Methylobacterium. Journal of Bacteriology, 184, 1818. |
| [25] | Mano H, Morisaki H (2008). Endophytic bacteria in the rice plant. Microbes and Environments, 23, 109-117. |
| [26] | Mano H, Tanaka F, Watanabe A, Kaga H, Okunishi S, Morisaki H (2006). Culturable surface and endophytic bacterial flora of the maturing seeds of rice plants (Oryza sativa) cultivated in a paddy field. Microbes and Environments, 21, 86-100. |
| [27] | McSpadden Gardener BB, Driks A (2004). Overview of the nature and application of biocontrol microbes: Bacillus spp. Phytopathology, 94, 1244. |
| [28] | Mullins TD, Britschgi TB, Krest RL, Giovannoni SJ (1995). Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnology and Oceanography, 40, 148-158. |
| [29] | Nelson EB (2004). Microbial dynamics and interactions in the spermosphere. Annual Review of Phytopathology, 42, 271-309. |
| [30] | Okunishi S, Sako K, Mano H, Imamura A, Morisaki H (2005). Bacterial flora of endophytes in the maturing seed of cultivated rice (Oryza sativa). Microbes and Environments, 20, 168-177. |
| [31] | Picard C, Bosco M (2006). Heterozygosis drives maize hybrids to select elite 2,4-diacethylphloroglucinol-producing Pseudomonas strains among resident soil populations. FEMS Microbiology Ecology, 58, 193-204. |
| [32] | Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH (1998). Bacillus thuringiensis and its pesticidal crystal proteins. Microbiology and Molecular Biology Reviews, 62, 775-806. |
| [33] | Senthilkumar M, Govindasamy V, Annapurna K (2007). Role of antibiosis in suppression of charcoal rot disease by soybean endophyte Paenibacillus sp. HKA-15. Current Microbiology, 55, 25-29. |
| [34] | Sessitsch A, Reiter B, Pfeifer U, Wilhelm E (2002). Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiology Ecology, 39, 23-32. |
| [35] | Shen DL (沈德龙), Feng YJ (冯永君), Song W (宋未) (2002). The effect of endophyte Pantoea agglomerans YS19 on the rice milk stage photosynthate distribution in flagleaf and fringe. Progress in Natural Science (自然科学进展), 12, 863-865. (in Chinese) |
| [36] | Soejima H, Sugiyama T, Ishihara K (1992). Changes in cytokinin activities and mass spectrometric analysis of cytokinins in root exudates of rice plant (Oryza sativa L.): comparison between cultivars Nipponbare and Akenohoshi. Plant Physiology, 100, 1724-1729. |
| [37] | Sturz AV, Matheson BG (1996). Populations of endophytic bacteria which influence host-resistance to Erwinia- induced bacterial soft rot in potato tubers. Plant and Soil, 184, 265-271. |
| [38] |
Sun L, Qiu FB, Zhang XX, Dai X, Dong XZ, Song W (2008). Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microbiology Ecology, 55, 415-424.
DOI URL |
| [39] | Trivedi P, Pandey A (2008). Plant growth promotion abilities and formulation of Bacillus megaterium strain B 388 (MTCC6521) isolated from a temperate Himalayan location. Indian Journal of Microbiology, 48, 342-347. |
| [40] | Ugoji EO, Laing MD, Hunter CH (2006). An investigation of the shelf-life (storage) of Bacillus isolates on seeds. South African Journal of Botany, 72, 28-33. |
| [41] | Vagelas IK, Pembroke B, Gowen SR, Davies KG (2007). The control of root-knot nematodes (Meloidogyne spp.) by Pseudomonas oryzihabitans and its immunological detection on tomato roots. Nematology, 9, 363-370. |
| [42] | van Overbeek L, van Elsas JD (2008). Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.). FEMS Microbiology Ecology, 64, 283-296. |
| [43] | Videira SS, de Araujo JLS, Rodrigues Lda S, Baldani VLD, Baldani JI (2009). Occurrence and diversity of nitrogen- fixing Sphingomonas bacteria associated with rice plants grown in Brazil. FEMS Microbiology Letters, 293, 11-19. |
| [44] | Völksch B, Thon S, Jacobsen ID, Gube M (2009). Polyphasic study of plant- and clinic-associated Pantoea agglomerans strains reveals indistinguishable virulence potential. Infection, Genetics and Evolution, 9, 1381-1391. |
| [45] | Weller DM (1988). Biological-control of soilborne plant- pathogens in the rhizosphere with bacteria. Annual Review of Phytopathology, 26, 379-407. |
| [46] | Xiao J, Li J, Yuan L, Tanksley SD (1995). Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics, 140, 745-754. |
| [47] | Xie ZW (谢中稳), Ge S (葛颂), Hong DY (洪德元) (1999). Preparation of DNA from silica gel dried mini-amount of leaves of Oryza rufipogon for RAPD study and total DNA bank construction. Acta Botanica Sinica (植物学报), 41, 807-812. (in Chinese with English abstract) |
| [48] | Yan H, Yu SH, Xie GL, Fang W, Su T, Li B (2010). Grain discoloration of rice caused by Pantoea ananatis (synonym Erwinia uredovora) in China. Plant Disease, 94, 482. |
| [49] | Yang HL (杨海莲), Sun XL (孙晓璐), Song W (宋未), Wang YS (王云山), Cai MY (蔡妙英) (1999). Screening, identification and distribution of endophytic associative diazotrophs isolated from rice plants. Acta Botanica Sinica (植物学报), 41, 927-931. (in Chinese with English abstract) |
| [1] | 朱湾湾, 王攀, 许艺馨, 李春环, 余海龙, 黄菊莹. 降水量变化与氮添加下荒漠草原土壤酶活性及其影响因素[J]. 植物生态学报, 2021, 45(3): 309-320. |
| [2] | 吴曼, 李娟娟, 刘金铭, 任安芝, 高玉葆. 刈割干扰和养分添加条件下Epichloë内生真菌感染对羽茅所在群落多样性和生产力的影响[J]. 植物生态学报, 2019, 43(2): 85-93. |
| [3] | 吴则焰,林文雄,陈志芳,方长旬,张志兴,吴林坤,周明明,陈婷. 中亚热带森林土壤微生物群落多样性随海拔梯度的变化[J]. 植物生态学报, 2013, 37(5): 397-406. |
| [4] | 梁存柱, 朱宗元, 王炜, 裴浩, 张韬, 王永利. 贺兰山植物群落类型多样性及其空间分异[J]. 植物生态学报, 2004, 28(3): 361-368. |
| [5] | 何田华, 葛颂. 植物种群交配系统、亲本分析以及基因流动研究[J]. 植物生态学报, 2001, 25(2): 144-154. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
Copyright © 2022 版权所有 《植物生态学报》编辑部
地址: 北京香山南辛村20号, 邮编: 100093
Tel.: 010-62836134, 62836138; Fax: 010-82599431; E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
备案号: 京ICP备16067583号-19