

Chin J Plant Ecol ›› 2023, Vol. 47 ›› Issue (12): 1629-1645.DOI: 10.17521/cjpe.2022.0295
• Research Articles • Previous Articles Next Articles
YANG Ling1,2,3,*, LIANG Si-Qi1,2,3,*, PAN Jia-Ming1,2,3, WEI Jin-Xin1,2,3, DING Tao4, JIANG Ri-Hong5, SHAO Yi-Zhen6, ZHANG Xian-Chun1,2, LIU Yong-Bo7,**( ), XIANG Qiao-Ping1,2,**(
), XIANG Qiao-Ping1,2,**( )
)
Received:2022-07-18
Accepted:2023-06-05
Online:2023-12-20
Published:2023-06-08
Contact:
**(Liu YB, liuyb@craes.org.cn; Xiang QP, qpxiang@ibcas.ac.cn)
About author:*Contributed equally to this work
Supported by:YANG Ling, LIANG Si-Qi, PAN Jia-Ming, WEI Jin-Xin, DING Tao, JIANG Ri-Hong, SHAO Yi-Zhen, ZHANG Xian-Chun, LIU Yong-Bo, XIANG Qiao-Ping. Species delimitation and genetic conservation of the endangered firs Abies beshanzuensis and A. ziyuanensis[J]. Chin J Plant Ecol, 2023, 47(12): 1629-1645.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.plant-ecology.com/EN/10.17521/cjpe.2022.0295
| 组群 Group | 种群 Population | 编号 Code | 数量 Number | 经度 Longitude (° E) | 纬度 Latitude (° N) | 海拔 Altitude (m) |
|---|---|---|---|---|---|---|
| 百山祖 Baishanzu (BSZ) | 浙江庆元县百山祖 Baishanzu, Qingyuan, Zhejiang | BSZ | 3 | 119.20 | 27.75 | 1 750 |
| 大院 Dayuan (DY) | 湖南炎陵县大院农场 Dayuan, Yanling, Hunan | DY | 3 | 114.07 | 26.45 | 1 478 |
| 江西井冈山市平水山 Pingshuishan, Jinggangshan, Jiangxi | JGS | 1 | 114.11 | 26.50 | 1 676 | |
| 广西全州县大云山 Dayunshan, Quanzhou, Guangxi | QZ | 3 | 110.96 | 26.38 | 1 801 | |
| 广西资源县银竹老山 Yinzhulaoshan, Ziyuan, Guangxi | YZ | 5 | 110.56 | 26.25 | 1 753 | |
| 资源 Ziyuan (ZY) | 湖南城步县金童山 Jintongshan, Chengbu, Hunan | CB | 4 | 110.58 | 26.28 | 1 646 |
| 湖南江永县千家峒 Qianjiadong, Jiangyong, Hunan | QJD | 3 | 111.30 | 25.57 | 1 800 | |
| 湖南新宁县舜皇山 Shunhuangshan, Xinning, Hunan | SHS | 1 | 110.56 | 26.22 | 1 778 |
Table 1 Geographic information and samples size of Abies beshanzuensis-A. ziyuanensis lineage
| 组群 Group | 种群 Population | 编号 Code | 数量 Number | 经度 Longitude (° E) | 纬度 Latitude (° N) | 海拔 Altitude (m) |
|---|---|---|---|---|---|---|
| 百山祖 Baishanzu (BSZ) | 浙江庆元县百山祖 Baishanzu, Qingyuan, Zhejiang | BSZ | 3 | 119.20 | 27.75 | 1 750 |
| 大院 Dayuan (DY) | 湖南炎陵县大院农场 Dayuan, Yanling, Hunan | DY | 3 | 114.07 | 26.45 | 1 478 |
| 江西井冈山市平水山 Pingshuishan, Jinggangshan, Jiangxi | JGS | 1 | 114.11 | 26.50 | 1 676 | |
| 广西全州县大云山 Dayunshan, Quanzhou, Guangxi | QZ | 3 | 110.96 | 26.38 | 1 801 | |
| 广西资源县银竹老山 Yinzhulaoshan, Ziyuan, Guangxi | YZ | 5 | 110.56 | 26.25 | 1 753 | |
| 资源 Ziyuan (ZY) | 湖南城步县金童山 Jintongshan, Chengbu, Hunan | CB | 4 | 110.58 | 26.28 | 1 646 |
| 湖南江永县千家峒 Qianjiadong, Jiangyong, Hunan | QJD | 3 | 111.30 | 25.57 | 1 800 | |
| 湖南新宁县舜皇山 Shunhuangshan, Xinning, Hunan | SHS | 1 | 110.56 | 26.22 | 1 778 |
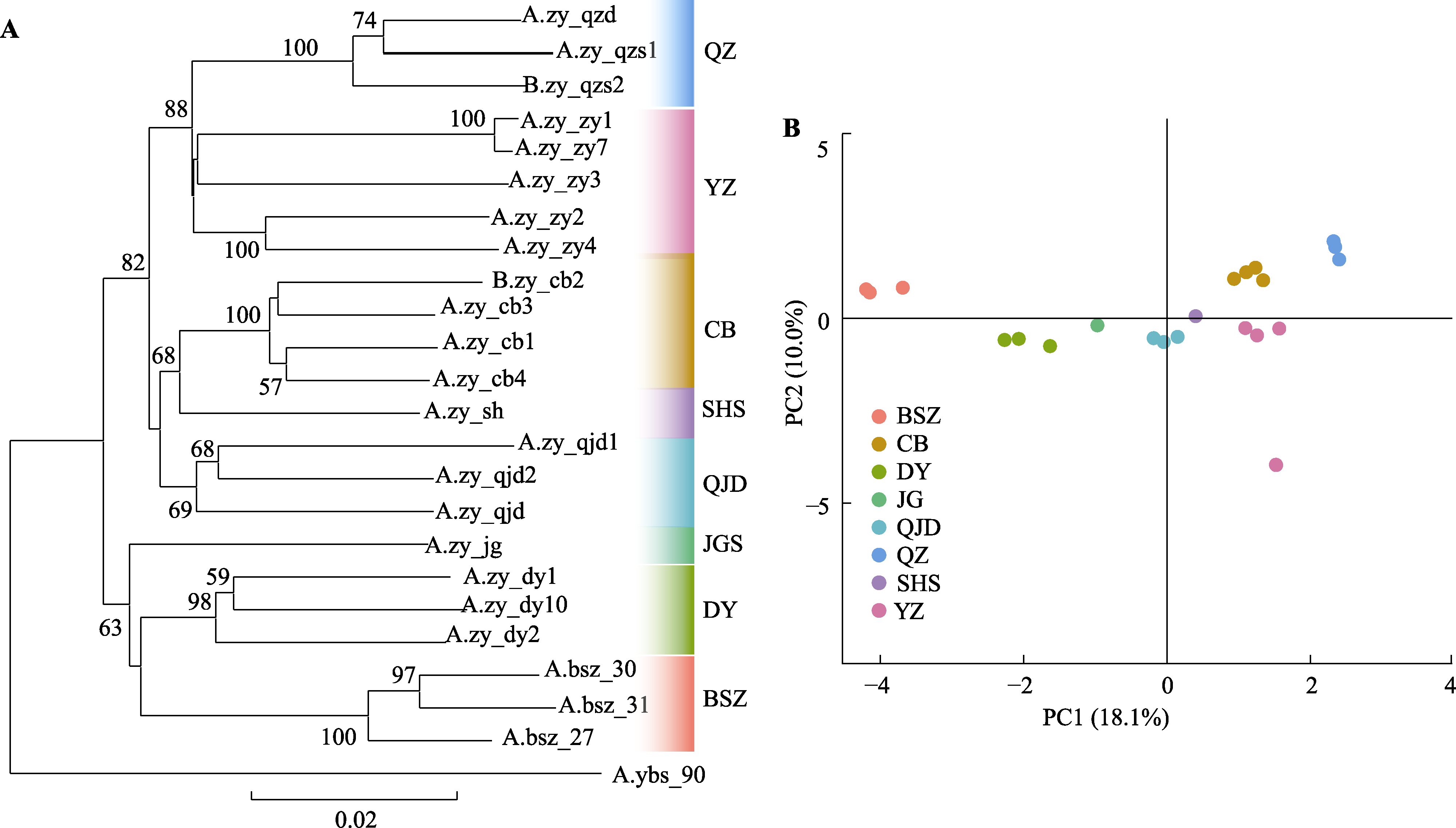
Fig. 1 Neighbor-joining phylogenetic tree (A) and PCA result (B) of Abies beshanzuensis-A. ziyuanensis lineage constructed using 805 nuclear single nucleotide polymorphisms. Numbers on branches are bootstrap value > 50% (1 000 replications). Population codes are shown in Table 1.
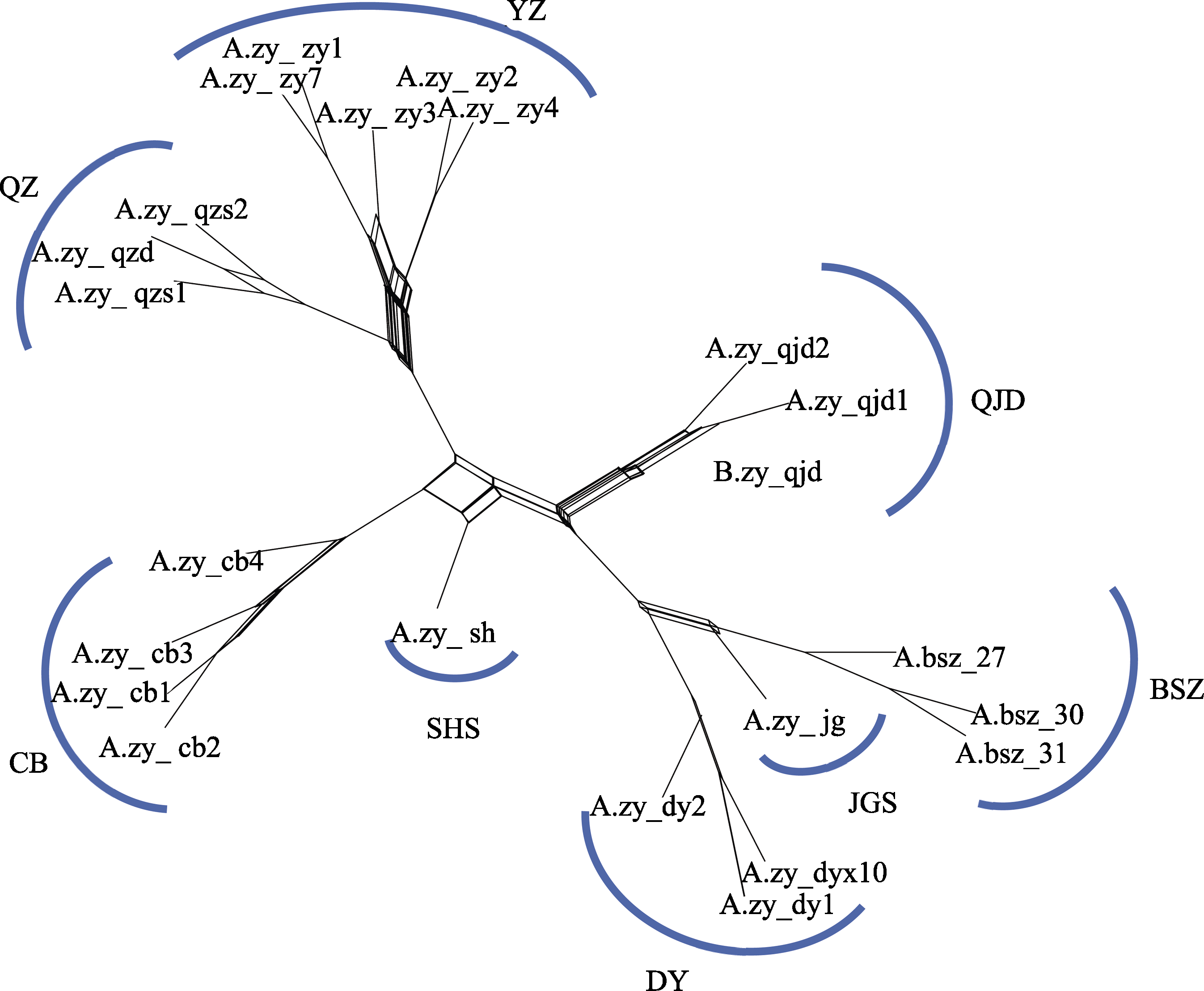
Fig. 2 Phylogenetic network of Abies beshanzuensis-A. ziyuanensis lineage based on 805 nuclear single nucleotide polymorphisms using SplitsTree. Population codes are shown in Table 1.
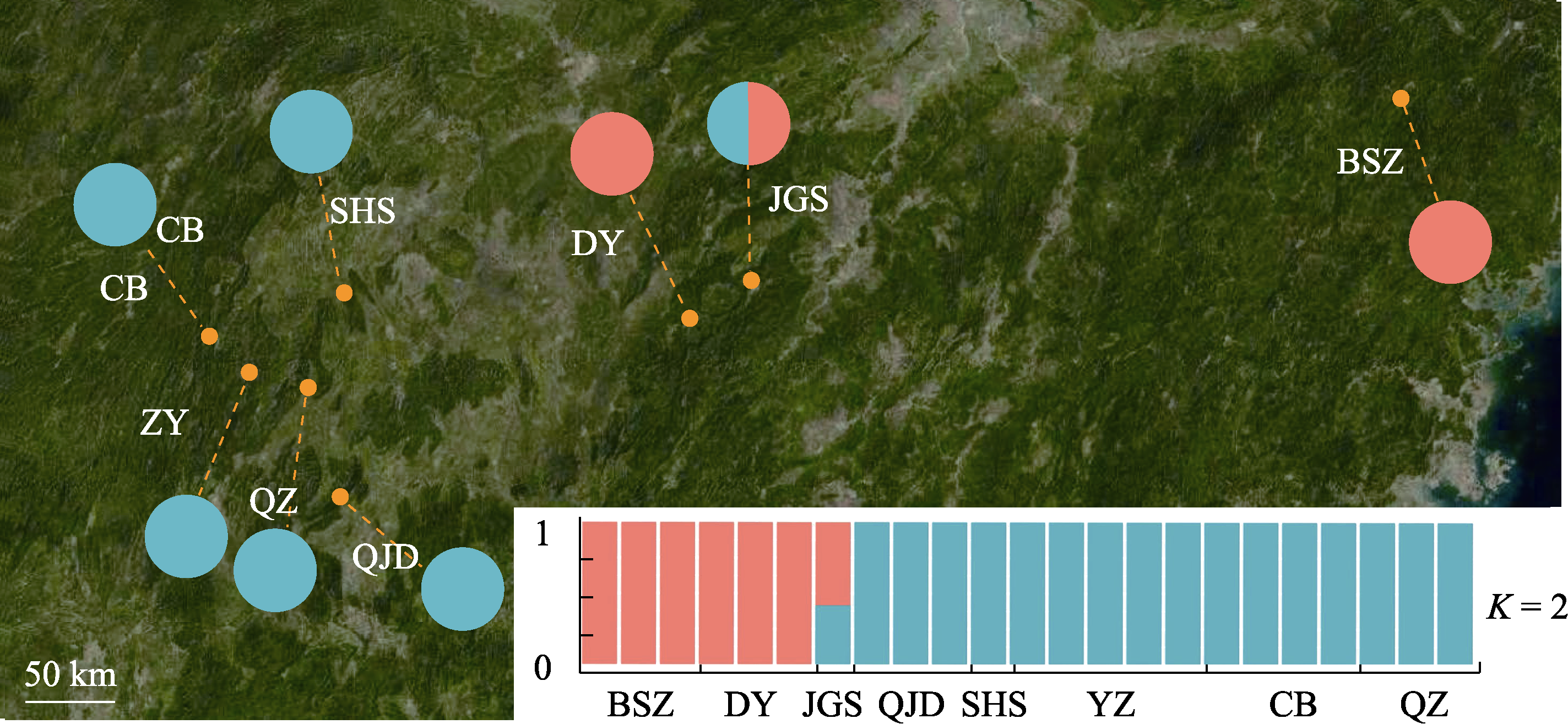
Fig. 3 Ancestral assignment of Abies beshanzuensis-A. ziyuanensis lineage based on 805 nuclear single nucleotide polymorphisms using ADMIXTURE (K = 2). Population codes are shown in Table 1.
| 种群 Population | 多态位点比例 Proportion of polymorphic markers | 核苷酸多样性 Nucleotide diversity | 期望杂合度 Expected heterozygosity | 观测杂合度 Observed heterozygosity | Tajima的D值 Tajima’s D |
|---|---|---|---|---|---|
| BSZ | 0.372 7 | 0.098 6 | 0.072 6 | 0.128 2 | 0.391 3 |
| DY | 0.444 7 | 0.115 3 | 0.063 0 | 0.104 3 | 0.307 4 |
| QJD | 0.438 5 | 0.108 0 | 0.066 6 | 0.099 1 | 0.185 1 |
| CB | 0.447 2 | 0.108 7 | 0.076 3 | 0.130 8 | 0.391 7 |
| ZY | 0.555 3 | 0.117 2 | 0.101 1 | 0.157 9 | 0.154 3 |
| QZ | 0.473 3 | 0.125 2 | 0.109 1 | 0.182 9 | 0.377 7 |
Table 2 Population genetic parameter of Abies beshanzuensis-A. ziyuanensis lineage
| 种群 Population | 多态位点比例 Proportion of polymorphic markers | 核苷酸多样性 Nucleotide diversity | 期望杂合度 Expected heterozygosity | 观测杂合度 Observed heterozygosity | Tajima的D值 Tajima’s D |
|---|---|---|---|---|---|
| BSZ | 0.372 7 | 0.098 6 | 0.072 6 | 0.128 2 | 0.391 3 |
| DY | 0.444 7 | 0.115 3 | 0.063 0 | 0.104 3 | 0.307 4 |
| QJD | 0.438 5 | 0.108 0 | 0.066 6 | 0.099 1 | 0.185 1 |
| CB | 0.447 2 | 0.108 7 | 0.076 3 | 0.130 8 | 0.391 7 |
| ZY | 0.555 3 | 0.117 2 | 0.101 1 | 0.157 9 | 0.154 3 |
| QZ | 0.473 3 | 0.125 2 | 0.109 1 | 0.182 9 | 0.377 7 |
| 变异来源 Source of variation | 自由度 df | 方差和 Sum of squares | 变异组分 Variance components | 占总变异比例 Percentage of variation (%) |
|---|---|---|---|---|
| 种群间 Among populations | 4 | 723.79 | 5.57 | 6.17 |
| 种群内 Within populations | 39 | 3 305.75 | 84.76 | 93.83 |
| 总计 Total | 45 | 4 029.54 | 93.18 |
Table 3 Results of analysis of molecular variance (AMOVA) from populations of Abies beshanzuensis-A. ziyuanensis lineage (p < 0.01)
| 变异来源 Source of variation | 自由度 df | 方差和 Sum of squares | 变异组分 Variance components | 占总变异比例 Percentage of variation (%) |
|---|---|---|---|---|
| 种群间 Among populations | 4 | 723.79 | 5.57 | 6.17 |
| 种群内 Within populations | 39 | 3 305.75 | 84.76 | 93.83 |
| 总计 Total | 45 | 4 029.54 | 93.18 |
| 种群 Population | BSZ | DY | QJD | CB | YZ | QZ |
|---|---|---|---|---|---|---|
| BSZ | - | |||||
| DY | 0.145 4 | - | ||||
| QJD | 0.140 2 | 0.079 9 | - | |||
| CB | 0.184 1 | 0.114 0 | 0.075 8 | - | ||
| YZ | 0.168 6 | 0.113 7 | 0.055 5 | 0.088 8 | - | |
| QZ | 0.207 5 | 0.169 1 | 0.110 1 | 0.126 3 | 0.083 1 | - |
Table 4 Fixation index (FST) between populations of Abies beshanzuensis-A. ziyuanensis lineage
| 种群 Population | BSZ | DY | QJD | CB | YZ | QZ |
|---|---|---|---|---|---|---|
| BSZ | - | |||||
| DY | 0.145 4 | - | ||||
| QJD | 0.140 2 | 0.079 9 | - | |||
| CB | 0.184 1 | 0.114 0 | 0.075 8 | - | ||
| YZ | 0.168 6 | 0.113 7 | 0.055 5 | 0.088 8 | - | |
| QZ | 0.207 5 | 0.169 1 | 0.110 1 | 0.126 3 | 0.083 1 | - |
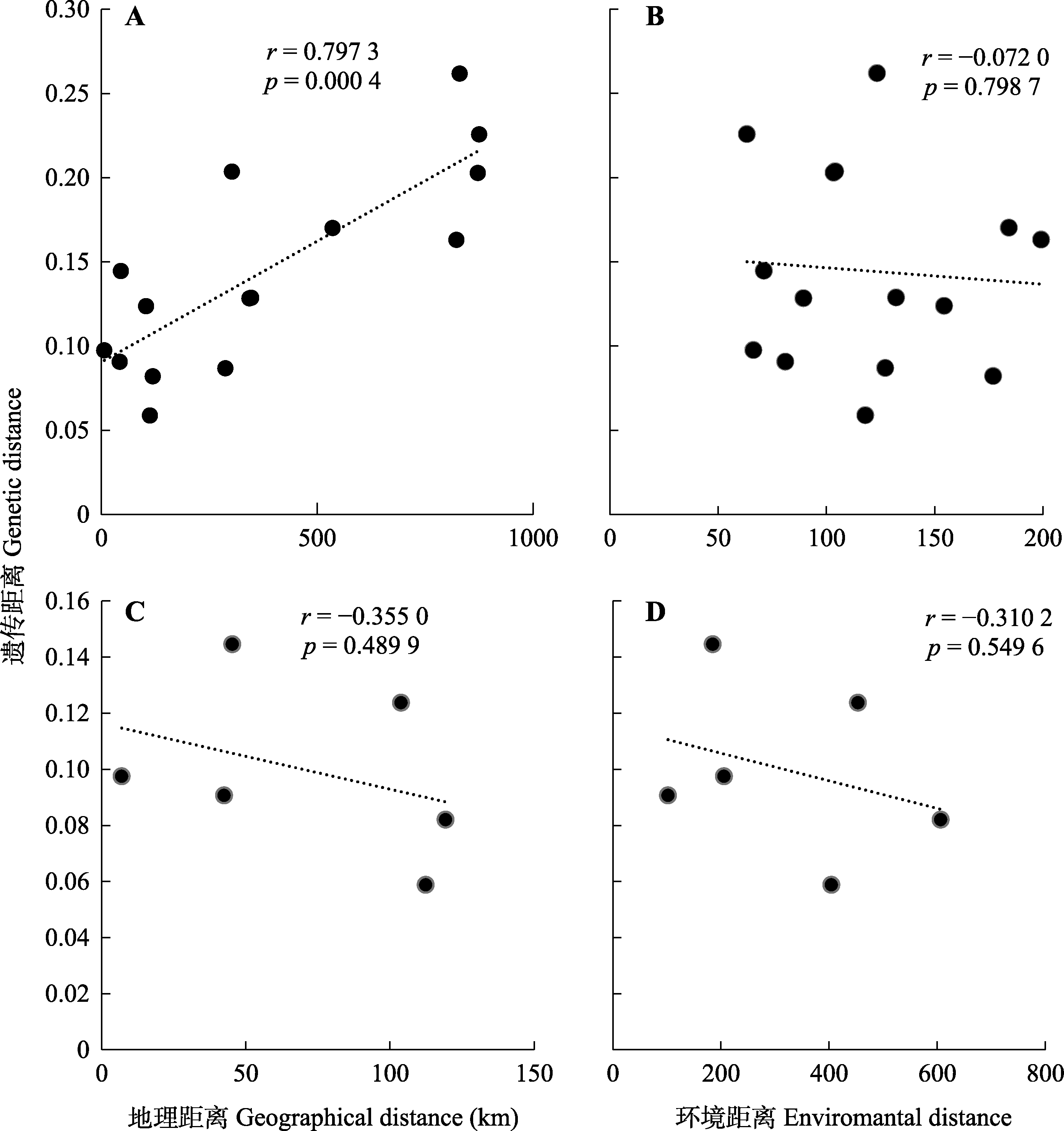
Fig. 4 Mantel test between genetic distance and geographic distance or environmental distance among populations of Abies beshanzuensis-A. ziyuanensis lineage (A, B) and A. ziyuanensis (C, D).
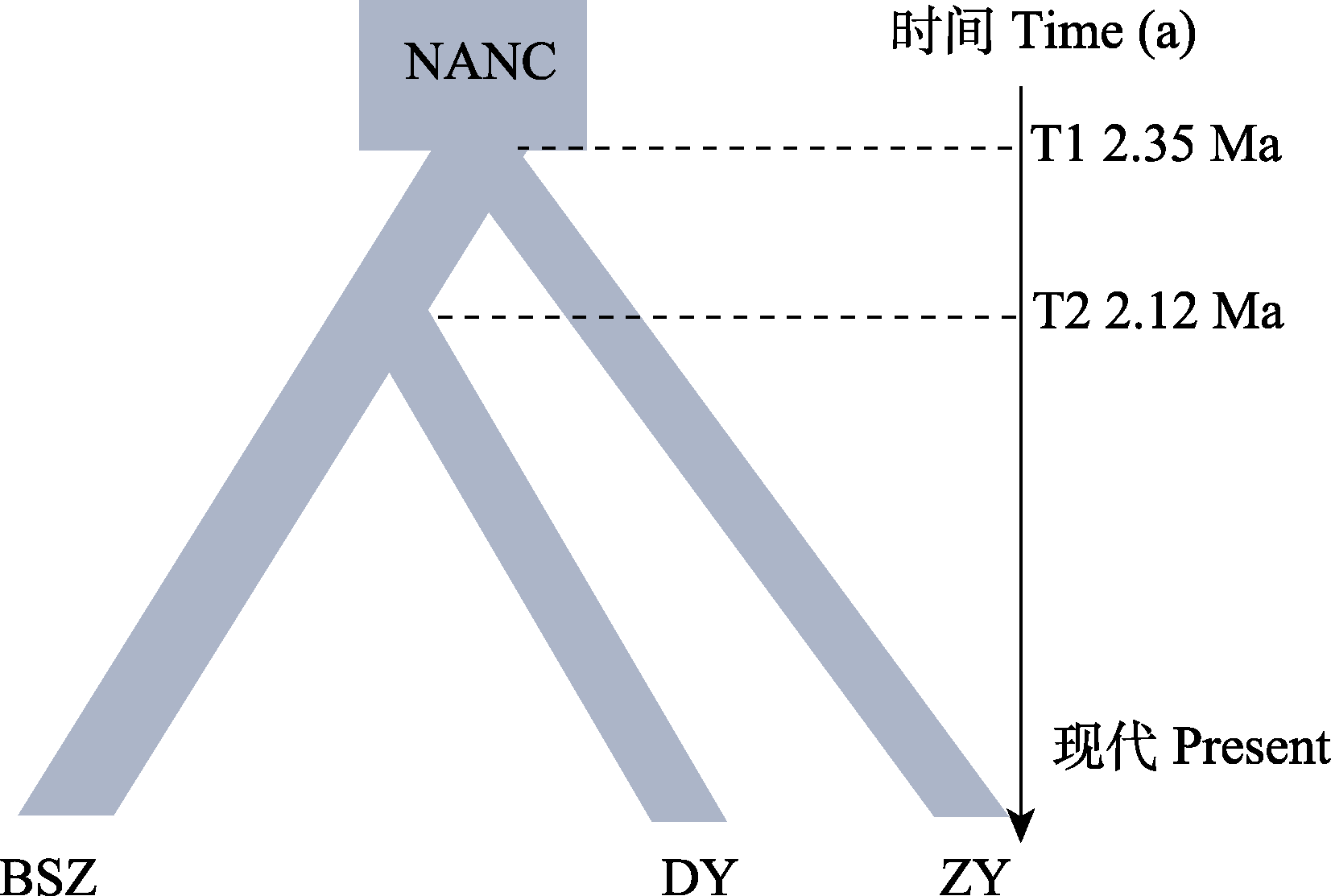
Fig. 5 The best demographic model of Abies beshanzuensis-A. ziyuanensis lineage. The width of the boxes represents the relative effective population size; T1 and T2 represent the first and second time of lineage differentiation, respectively; NANC indicates ancestral population size; BSZ represents the Baishanzu group; DY represents the Dayuan group; ZY represents the Ziyuan group.
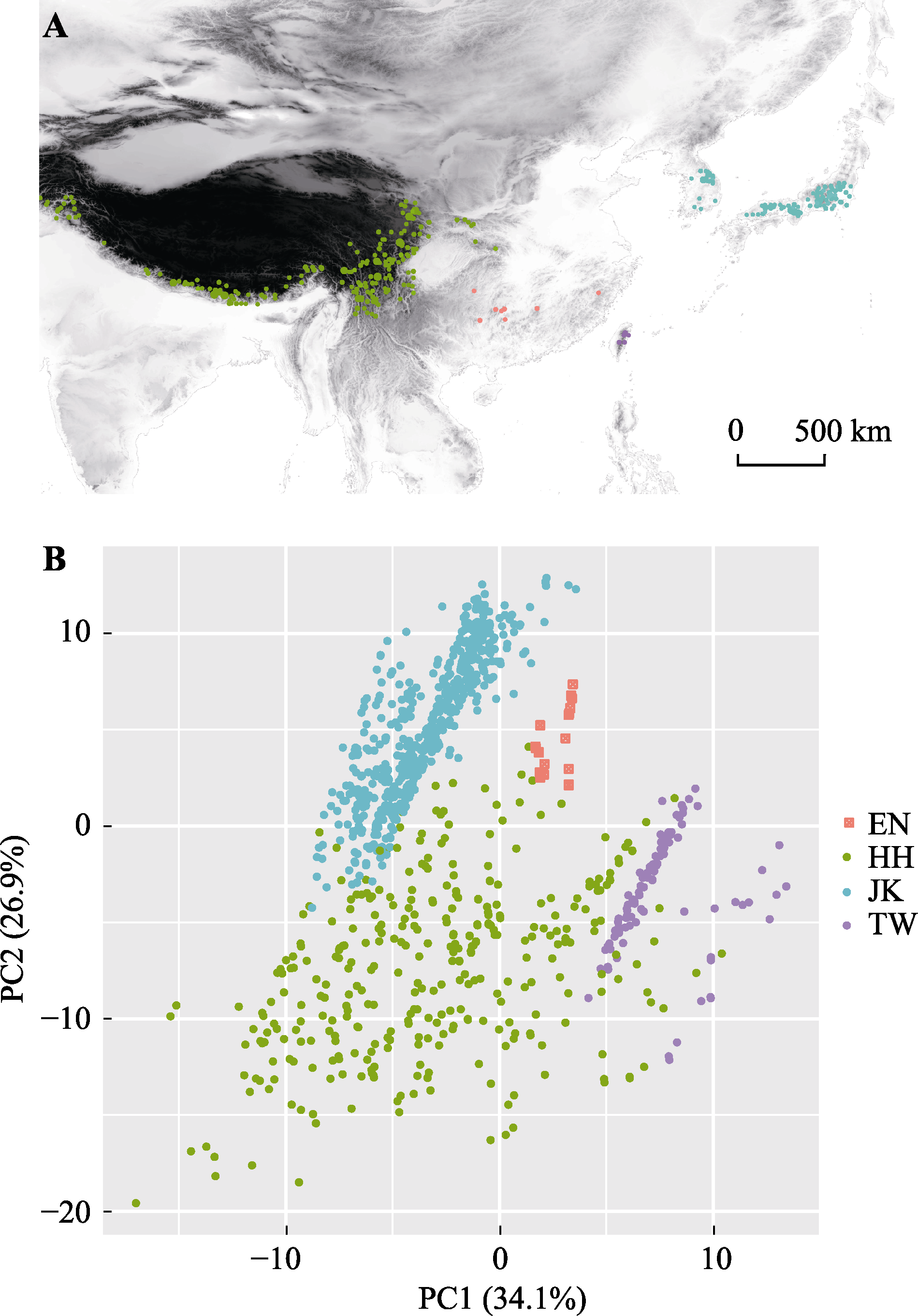
Fig. 6 Geographical distribution map (A) and principal component (PC) analysis of 19 climatic factors (B) of firs in East Asia. EN, endangered firs distributed in subtropical mountains of China; HH, firs in Hengduan-Himalayan mountains; JK, firs in Japan-Korean peninsula; TW, firs in Taiwan mountains of China.
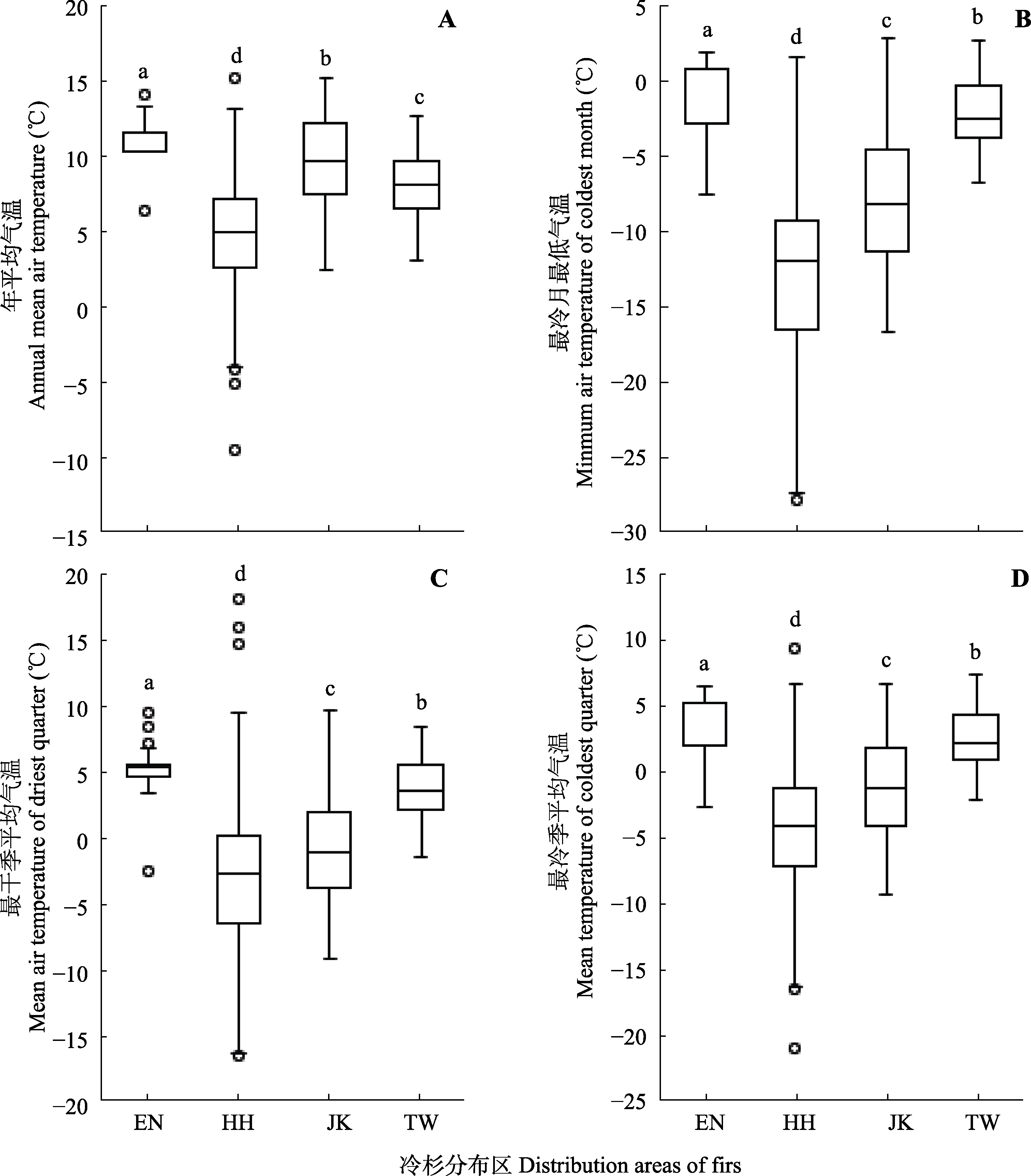
Fig. 7 Results of ANOVA and multiple comparative analysis for Abies in East Asia. EN, endangered firs distributed in subtropical mountains of China; HH, firs in Hengduan-Himalayan mountains; JK, firs in Japan-Korean peninsula; TW, firs in Taiwan mountains of China. Different lowercase letters indicate significant difference (p < 0.01).
| 物种 Species | 冬芽 Winter bud | 球果 Seed cones | 种翅 Seed wings | 地理分布 Distribution | 海拔 Altitude (m) | |||
|---|---|---|---|---|---|---|---|---|
| 形状 Shape | 成熟时颜色 Color when ripe | 苞鳞 Bract scales | 中部种鳞 Middle seed scales | |||||
| 百山祖冷杉 A. beshanzuensis | 卵圆形至圆锥形 Ovoid to conical | 圆柱形 Cylindric | 淡褐色或淡褐黄色 Light brown or brownish yellow | 苞鳞上部宽7-8 mm, 中下部渐窄, 宽3-4 mm Bract 7-8 mm wide at upper part, middle and lower part gradually attenuate, 3-4 mm wide | 扇状四边形或肾状四边形, 长1.8-2.4 cm, 宽2.5- 3 cm Flabellate- trapeziform or reniform- trapeziform, 1.8-2.4 cm × 2.5- 3 cm | 种子(含种翅)长1.0-2.3 cm, 种翅与种子等长 Length of seed wing as long as the seed, including seed 1.0-2.3 cm | 浙江(庆元县百山祖)、湖南(炎陵县大院农场)、江西(井冈山市平水山、遂川县南风面) Zhejiang: Qingyuan (Baishanzu), Hunan: Yanling (Dayuan); Jiangxi: Jinggangshan (Pingshuishan), Suichuan (Nanfengmian) | 1 350-1 950 |
| 资源冷杉 A. ziyuanensis | 圆锥形或圆锥状卵圆形 Conical or conical-ovoid | 长椭圆状圆柱形 Long ellipsoid-cylindric | 暗绿褐色或暗褐色 Dark greenish brown or dark brown | 苞鳞上部宽9-10 mm, 中部及中下部明显变窄, 宽约3 mm Bract 9-10 mm wide at upper part, middle and lower part conspicuous attenuate, 3 mm wide | 扇状四边形, 长2.3-2.5 cm, 宽3- 3.3 cm Flabellate- trapeziform, 2.3-2.5 cm × 3-3.3 cm | 种子(含种翅)长2.4-3.1 cm, 种翅较种子长一倍以上 Length of seed wing two times that of the seed, including seed 2.4-3.1 cm | 广西(资源县银竹老山、全州县)、湖南(新宁县舜皇山、城步县金童山、江永县千家峒) Guangxi: Ziyuan (Yinzhulaoshan), Quanzhou; Hunan: Xinning (Shunhuangshan), Chengbu (Jintongshan), Jiangyong (Qianjiadong) | 1 600-1 800 |
Table 5 Morphological characteristics and geographical distribution of Abies beshanzuensis and A. ziyuanensis
| 物种 Species | 冬芽 Winter bud | 球果 Seed cones | 种翅 Seed wings | 地理分布 Distribution | 海拔 Altitude (m) | |||
|---|---|---|---|---|---|---|---|---|
| 形状 Shape | 成熟时颜色 Color when ripe | 苞鳞 Bract scales | 中部种鳞 Middle seed scales | |||||
| 百山祖冷杉 A. beshanzuensis | 卵圆形至圆锥形 Ovoid to conical | 圆柱形 Cylindric | 淡褐色或淡褐黄色 Light brown or brownish yellow | 苞鳞上部宽7-8 mm, 中下部渐窄, 宽3-4 mm Bract 7-8 mm wide at upper part, middle and lower part gradually attenuate, 3-4 mm wide | 扇状四边形或肾状四边形, 长1.8-2.4 cm, 宽2.5- 3 cm Flabellate- trapeziform or reniform- trapeziform, 1.8-2.4 cm × 2.5- 3 cm | 种子(含种翅)长1.0-2.3 cm, 种翅与种子等长 Length of seed wing as long as the seed, including seed 1.0-2.3 cm | 浙江(庆元县百山祖)、湖南(炎陵县大院农场)、江西(井冈山市平水山、遂川县南风面) Zhejiang: Qingyuan (Baishanzu), Hunan: Yanling (Dayuan); Jiangxi: Jinggangshan (Pingshuishan), Suichuan (Nanfengmian) | 1 350-1 950 |
| 资源冷杉 A. ziyuanensis | 圆锥形或圆锥状卵圆形 Conical or conical-ovoid | 长椭圆状圆柱形 Long ellipsoid-cylindric | 暗绿褐色或暗褐色 Dark greenish brown or dark brown | 苞鳞上部宽9-10 mm, 中部及中下部明显变窄, 宽约3 mm Bract 9-10 mm wide at upper part, middle and lower part conspicuous attenuate, 3 mm wide | 扇状四边形, 长2.3-2.5 cm, 宽3- 3.3 cm Flabellate- trapeziform, 2.3-2.5 cm × 3-3.3 cm | 种子(含种翅)长2.4-3.1 cm, 种翅较种子长一倍以上 Length of seed wing two times that of the seed, including seed 2.4-3.1 cm | 广西(资源县银竹老山、全州县)、湖南(新宁县舜皇山、城步县金童山、江永县千家峒) Guangxi: Ziyuan (Yinzhulaoshan), Quanzhou; Hunan: Xinning (Shunhuangshan), Chengbu (Jintongshan), Jiangyong (Qianjiadong) | 1 600-1 800 |
| [1] |
Alexander DH, Novembre J, Lange K (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Research, 19, 1655-1664.
DOI PMID |
| [2] |
Azaiez A, Pavy N, Gérardi S, Laroche J, Boyle B, Gagnon F, Mottet MJ, Beaulieu J, Bousquet J (2018). A catalog of annotated high-confidence SNPs from exome capture and sequencing reveals highly polymorphic genes in Norway spruce (Picea abies). BMC Genomics, 19, 942. DOI: 10.1186/s12864-018-5247-z.
PMID |
| [3] |
Capblancq T, Butnor JR, Deyoung S, Thibault E, Munson H, Nelson DM, Fitzpatrick MC, Keller SR (2020). Whole- exome sequencing reveals a long-term decline in effective population size of red spruce (Picea rubens). Evolutionary Applications, 13, 2190-2205.
DOI PMID |
| [4] |
Chen J, Li L, Milesi P, Jansson G, Berlin M, Karlsson B, Aleksic J, Vendramin GG, Lascoux M (2019). Genomic data provide new insights on the demographic history and the extent of recent material transfers in Norway spruce. Evolutionary Applications, 12, 1539-1551.
DOI PMID |
| [5] | Chen SF, Zhou YQ, Chen YR, Gu J (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics, 34, i884-i890. |
| [6] | Dai WJ, Tang SQ, Liu YH (2006). Study on genetic diversity of the endangered plant Abies ziyuanensis by using chloroplast SSR marker. Guangxi Sciences, 13, 151-155. |
| [ 代文娟, 唐绍清, 刘燕华 (2006). 叶绿体微卫星分析濒危植物资源冷杉的遗传多样性. 广西科学, 13, 151-155.] | |
| [7] |
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R,1000 Genomes Project Analysis Group (2011). The variant call format and VCFtools. Bioinformatics, 27, 2156-2158.
DOI PMID |
| [8] |
Davis MB, Shaw RG (2001). Range shifts and adaptive responses to Quaternary climate change. Science, 292, 673-679.
DOI PMID |
| [9] |
de la Torre AR, Li Z, Van de Peer Y, Ingvarsson PK (2017). Contrasting rates of molecular evolution and patterns of selection among gymnosperms and flowering plants. Molecular Biology and Evolution, 34, 1363-1377.
DOI PMID |
| [10] | Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M (2013). Robust demographic inference from genomic and SNP data. PLoS Genetics, 9, e1003905. DOI: 10.1371/ journal.pgen.1003905. |
| [11] |
Excoffier L, Lischer HEL (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567.
DOI PMID |
| [12] |
Excoffier L, Marchi N, Marques DA, Matthey-Doret R, Gouy A, Sousa VC (2021). Fastsimcoal2: demographic inference under complex evolutionary scenarios. Bioinformatics, 37, 4882-4885.
DOI PMID |
| [13] | Feng L, Du FK (2022). Landscape genomics in tree conservation under a changing environment. Frontiers in Plant Science, 13, 822217. DOI: 10.3389/fpls.2022.822217. |
| [14] |
Foll M, Gaggiotti O (2008). A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics, 180, 977-993.
DOI PMID |
| [15] | Fu L, Li N, Elias TS (1999). Abies//Wu Z, Raven PH. Flora of China: Vol. 4. Science Press & Missouri Botanical Garden Press, Beijing & St. Louis. 44-52. |
| [16] | Fu LG, Lü YJ, Mo XL (1980). The genus Abies discovered for the first time in Guangxi and Hunan. Acta Phytotaxonomica Sinica, 18, 205-210. |
| [ 傅立国, 吕庸浚, 莫新礼 (1980). 冷杉属植物在广西和湖南首次发现. 植物分类学报, 18, 205-210.] | |
| [17] | Furlan E, Stoklosa J, Griffiths J, Gust N, Ellis R, Huggins RM, Weeks AR (2012). Small population size and extremely low levels of genetic diversity in island populations of the platypus, Ornithorhynchus anatinus. Ecology and Evolution, 2, 844-857. |
| [18] |
Gernandt DS, Aguirre Dugua X, Vázquez-Lobo A, Willyard A, Moreno Letelier A, Pérez de la Rosa JA, Piñero D, Liston A (2018). Multi-locus phylogenetics, lineage sorting, and reticulation in Pinus subsection Australes. American Journal of Botany, 105, 711-725.
DOI PMID |
| [19] | Hamrick JL, Godt MJW (1992). Allozyme diversity in plant systematics//Soltis PS, Soltis DE, Doyle JJ. Molecular Systematic of Plants. Chapman and Hill, New York. 54. |
| [20] |
Hewitt G (2000). The genetic legacy of the Quaternary ice ages. Nature, 405, 907-913.
DOI |
| [21] |
Huson DH, Bryant D (2006). Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution, 23, 254-267.
DOI PMID |
| [22] | International Union for Conservation of Nature and Natural Resources (IUCN) (2019). Guidelines for using the IUCN red list categories and criteria, version 14. [2020-12-24]. https://www.iucnredlist.org/documents/RedListGuidelines.pdf. |
| [23] | Johnson MG, Gardner EM, Liu Y, Medina R, Goffinet B, Shaw AJ, Zerega NJC, Wickett NJ (2016). HybPiper: extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Applications in Plant Sciences, 4, 1600016. DOI: 10.3732/ apps.1600016. |
| [24] |
Jombart T (2008). Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics, 24, 1403-1405.
DOI PMID |
| [25] | Li GQ (2011). Evaluation the Ecological Niche Models and Predicting Species Potential Distribution Area. PhD dissertation, Institute of Botany, the Chinese Academy of Sciences, Beijing. |
| [ 李国庆 (2011). 物种生态位模型的适用性评价和物种潜在分布区预测. 博士学位论文, 中国科学院植物研究所, 北京.] | |
| [26] |
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R,Genome Project Data Processing Subgroup 1000 (2009). The sequence alignment/ map format and SAMtools. Bioinformatics, 25, 2078-2079.
DOI URL |
| [27] |
Li N, Fu LK (1997). Notes on gymnosperms I. Taxonomic treatments of some Chinese conifers. Novon, 7, 261-264.
DOI URL |
| [28] |
Liu J, Möller M, Provan J, Gao L, Poudel RC, Li D (2013). Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot. New Phytologist, 199, 1093-1108.
DOI PMID |
| [29] |
Liu JQ (2016). “The integrative species concept” and “species on the speciation way”. Biodiversity Science, 24, 1004- 1008.
DOI URL |
|
[ 刘建全 (2016). “整合物种概念”和“分化路上的物种”. 生物多样性, 24, 1004-1008.]
DOI |
|
| [30] | Liu L, Hao ZZ, Liu YY, Wei XX, Cun YZ, Wang XQ (2014). Phylogeography of Pinus armandii and its relatives: heterogeneous contributions of geography and climate changes to the genetic differentiation and diversification of Chinese white pines. PLoS ONE, 9, e85920. DOI: 10.1371/journal.pone.0085920. |
| [31] | Liu QX (1982). The discovery, function and significance of Abies dayuanensis. Hunan Forestry Science & Technology, (3), 10-17. |
| [ 刘起衔 (1982). 大院冷杉的发现及其作用和意义. 湖南林业科技, (3), 10-17.] | |
| [32] | Liu QX (1988). New plants from Hunan. Bulletin of Botancal Research, 8(3), 85-94. |
| [ 刘起衔 (1988). 湖南产新植物. 植物研究, 8(3), 85-94.] | |
| [33] | Liu R, Wang CJ, He J, Zhang ZX (2018). Analysis of geographical distribution of Abies in China under climate change. Bulletin of Botanical Research, 38, 37-46. |
|
[ 刘然, 王春晶, 何健, 张志翔 (2018). 气候变化背景下中国冷杉属植物地理分布模拟分析. 植物研究, 38, 37-46.]
DOI |
|
| [34] |
Mastretta-Yanes A, Moreno-Letelier A, Piñero D, Jorgensen TH, Emerson BC (2015). Biodiversity in the Mexican highlands and the interaction of geology, geography and climate within the Trans-Mexican Volcanic Belt. Journal of Biogeography, 42, 1586-1600.
DOI URL |
| [35] |
Meirmans PG (2012). The trouble with isolation by distance. Molecular Ecology, 21, 2839-2846.
DOI PMID |
| [36] |
Melo FPL, Arroyo-Rodríguez V, Fahrig L, Martínez-Ramos M, Tabarelli M (2013). On the hope for biodiversity-friendly tropical landscapes. Trends in Ecology & Evolution, 28, 462-468.
DOI URL |
| [37] |
Peakall R, Smouse PE (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics, 28, 2537-2539.
PMID |
| [38] |
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, Daly MJ, Sham PC (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics, 81, 559-575.
DOI PMID |
| [39] | Qin A, Ding Y, Jian Z, Ma F, Worth JRP, Pei S, Xu G, Guo Q, Shi Z (2021). Low genetic diversity and population differentiation in Thuja sutchuenensis Franch., an extremely endangered rediscovered conifer species in southwestern China. Global Ecology and Conservation, 25, e01430. DOI: 10.1016/j.gecco.2020.e01430. |
| [40] |
Qiu Y, Fu C, Comes HP (2011). Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Molecular Phylogenetics and Evolution, 59, 225-244.
DOI URL |
| [41] |
Qiu ZW, Jiang HE, Ding LL, Shang X (2020). Late Pleistocene- Holocene vegetation history and anthropogenic activities deduced from pollen spectra and archaeological data at Guxu Lake, eastern China. Scientific Reports, 10, 9306. DIO: 10.1038/s41598-020-65834-z.
DOI |
| [42] | Shao YZ, Yuan ZL, Liu YY, Liu FQ, Xiang RC, Zhang YY, Ye YZ, Chen Y, Wen Q (2022). Glacial expansion or interglacial expansion? contrasting demographic models of four cold-adapted fir species in North Americaand East Asia. Frontiers in Ecology and Evolution, 10, 844354. DOI: 10.3389/FEVO.2022.844354. |
| [43] | Shao Y, Zhang X, Phan LK, Xiang Q (2017). Elevation shift in Abies Mill. (Pinaceae) of subtropical and temperate China and vietnam-corroborative evidence from cytoplasmic DNA and ecological niche modeling. Frontiers in Plant Science, 8, 578. DOI: 10.3389/fpls.2017.00578. |
| [44] | Su HL, Tang SQ (2004). Genetic diversity of the endangered plant Abies ziyuanensis in two populations. Guihaia, 24, 414-417. |
| [ 苏何玲, 唐绍清 (2004). 濒危植物资源冷杉遗传多样性研究. 广西植物, 24, 414-417.] | |
| [45] |
Su JY, Yan Y, Li C, Li D, Du F (2020). Informing conservation strategies with genetic diversity in wild plant with extremely small populations: a review on gymnosperms. Biodiversity Science, 28, 376-384.
DOI |
|
[ 苏金源, 燕语, 李冲, 李丹, 杜芳 (2020). 通过遗传多样性探讨极小群体野生植物的致濒机理及保护策略: 以裸子植物为例. 生物多样性, 28, 376-384.]
DOI |
|
| [46] |
Tsumura Y, Yoshimura K, Tomaru N, Ohba K (1995). Molecular phytogeny of conifers using RFLP analysis of PCR-amplified specific chloroplast genes. Theoretical and Applied Genetics, 91, 1222-1236.
DOI PMID |
| [47] | Turland NJ, Wiersema JH, Barrie FR, Greuter W, Hawksworth DL, Herendeen PS, Knapp S, Kusber WH, Li D, Marhold K, May TW, Mcneill J, Monro AM, Prado J, Price MJ, Smith GF (2018). International code of nomenclature for algae, fungi, and plants (Shenzhen code). Regnum Vegetabile. Koeltz Botanical Books, Glashütten. 159. |
| [48] | van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA (2013). From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Current Protocols in Bioinformatics, 43, 1110. DOI: 10.1002/0471250953.bi1110s43. |
| [49] |
Wang J, Abbott RJ, Peng Y, Du F, Liu J (2011). Species delimitation and biogeography of two fir species (Abies) in central China: cytoplasmic DNA variation. Heredity, 107, 362-370.
DOI PMID |
| [50] | Wang QC, Li H, Li XX (2012). Geographical distribution characters and generating mechanisms of genus Abies in China. Journal of Central South University of Forestry & Technology, 32(9), 11-15. |
| [ 王清春, 李晖, 李晓笑 (2012). 中国冷杉属植物的地理分布特征及成因初探. 中南林业科技大学学报, 32(9), 11-15.] | |
| [51] | Wei F, Tang SQ (2011). Genetic diversity of endangered plant Abies ziyuanensis based on nrDNA GapC gene intron sequences. Bulletin of Botanical Research, 31, 729-734. |
|
[ 韦范, 唐绍清 (2011). 基于nrDNA GapC基因内含子序列的资源冷杉遗传多样性研究. 植物研究, 31, 729-734.]
DOI |
|
| [52] | Wu MX (1976). Abies beshanzuensis M. H. Wu—A new species of Abies from Chekiang. Acta Phytotaxonomica Sinica, 14, 15-21. |
| [ 吴鸣翔 (1976). 百山祖冷杉——一种新的冷杉的发现. 植物分类学报, 14, 15-21.] | |
| [53] | Wu YG, Rao LB, Chen DL, Zhou RF, Ye ZL (2010). Artificial seedling-raising of Abies beshanzuensis seed. Journal of Anhui Agricultural Sciences, 38, 12038-12039. |
| [ 吴友贵, 饶龙兵, 陈德良, 周荣飞, 叶珍林 (2010). 百山祖冷杉种子的人工育苗试验. 安徽农业科学, 38, 12038-12039.] | |
| [54] | Wu YG, Zhou RF, Chen XR, Ye ZL, Xia JT (2009). Comparison of the growth of Abies beshanzuensis under different cultivated environments. Forestry Science & Technology, 34(6), 8-10. |
| [ 吴友贵, 周荣飞, 陈小荣, 叶珍林, 夏家天 (2009). 百山祖冷杉小苗在不同生境下的生长比较研究. 林业科技, 34(6), 8-10.] | |
| [55] | Xiang QP (2001). A preliminary survey on the distribution of rare and endangered plants of Abies in China. Guihaia, 21, 113-117. |
| [ 向巧萍 (2001). 中国的几种珍稀濒危冷杉属植物及其地理分布成因的探讨. 广西植物, 21, 113-117.] | |
| [56] | Xiang XG, Cao M, Zhou ZK (2006). Fossil history and modern distribution of the genus Abies (Pinaceae). Acta Botanica Yunnanica, 28, 439-452. |
| [ 向小果, 曹明, 周浙昆 (2006). 松科冷杉属植物的化石历史和现代分布. 云南植物研究, 28, 439-452.] | |
| [57] | Yan HF, Zhang CY, Wang FY, Hu CM, Ge XJ, Hao G (2012). Population expanding with the phalanx model and lineages split by environmental heterogeneity: a case study of Primula obconica in subtropical China. PLoS ONE, 7, e41315. DOI: 10.1371/journal.pone.0041315. |
| [58] | Yuan H, Atta C, Tornabene L, Li C (2019). Assexon: assembling exon using gene capture data. Evolutionary Bioinformatics Online, 15, 1176934319874792. DOI: 10.1177/1176934319874792. |
| [59] |
Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li W, Wang G (2020). PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 20, 348-355.
DOI PMID |
| [60] | Zhang DY, Jiang XH (1999). Progress in studies of genetic diversity and conservation biology of endangered plant species. Chinese Biodiversity, 7, 31-37. |
| [ 张大勇, 姜新华 (1999). 遗传多样性与濒危植物保护生物学研究进展. 生物多样性, 7, 31-37.] | |
| [61] | Zhang YR (2009). Studies on Endangered Mechanism and Population Conservation of Abies ziyuanensis. PhD dissertation, Beijing Forestry University, Beijing. |
| [ 张玉荣 (2009). 资源冷杉的濒危机制与群体保育研究. 博士学位论文, 北京林业大学, 北京.] | |
| [62] | Zhou ZG, Wen X, Zhong PH, Xiao ZY (2020). Study on the population age structure and seedling growth of Abies beshanzuensis var. ziyuanensis in Mount Nanfengmian of Suichuan County. South China Forestry Science, 48(6), 35-39. |
| [ 周志光, 温馨, 钟平华, 肖忠优 (2020). 遂川南风面资源冷杉种群年龄结构及幼苗生长研究. 南方林业科学, 48(6), 35-39.] |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Chinese Journal of Plant Ecology
Tel: 010-62836134, 62836138, E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn