

Chin J Plant Ecol ›› 2020, Vol. 44 ›› Issue (6): 661-668.DOI: 10.17521/cjpe.2019.0298
• Research Articles • Previous Articles Next Articles
WANG Chun-Cheng1, MA Song-Mei2,*( ), ZHANG Dan1, WANG Shao-Ming1
), ZHANG Dan1, WANG Shao-Ming1
Received:2019-11-04
Accepted:2020-03-27
Online:2020-06-20
Published:2020-04-30
Contact:
MA Song-Mei: ORCID:0000-0002-3107-2256, shzmsm@126.com
Supported by:WANG Chun-Cheng, MA Song-Mei, ZHANG Dan, WANG Shao-Ming. Spatial genetic structure of Lycium ruthenicum in the Qaidam Basin[J]. Chin J Plant Ecol, 2020, 44(6): 661-668.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.plant-ecology.com/EN/10.17521/cjpe.2019.0298
| 种群名称及编码 Population name and code | 海拔 Altitude (m) | 经纬度 Latitude and Longitude | 采样数 Sample size | 单倍型及个体数 Haplotype and the individual numbers | 单倍型多样性 Haplotype diversity (Hd ± SD) | 核苷酸多样性 Nucleotide diversity (π ± SD) | |
|---|---|---|---|---|---|---|---|
| 德令哈市红光村 Hongguang Village of Delingha City (DLH) | 2 970 | 37.38° N, 97.34° E | 10 | H1 (2), H4 (8) | 0.356 ± 0.025 | 0.000 73 ± 0.33 | |
| 都兰县诺木洪林业站 Nuomuhong Forestry Station of Dulan County (NMH1) | 2 820 | 36.41° N, 96.45° E | 15 | H5 (7), H6 (6), H7 (2) | 0.604 ± 0.069 | 0.000 46 ± 0.09 | |
| 都兰县诺木洪乡田格力村 Tiangeli Village of Nuomuhong Township of Dulan County (NMH2) | 2 762 | 36.39° N, 96.19° E | 12 | H5 (5), H7 (7) | 0.530 ± 0.053 | 0.000 37 ± 0.05 | |
| 格尔木市新乐村 Xinle Village of Golmud City (GEM1) | 2 787 | 36.39° N, 94.86° E | 12 | H1 (7), H2 (4), H3 (1) | 0.591 ± 0.011 | 0.001 26 ± 0.03 | |
| 格尔木市大格勒乡 Dagele Township of Golmud City (GEM2) | 2 837 | 36.44° N, 95.75° E | 11 | H1 (3), H2 (8) | 0.436 ± 0.018 | 0.000 09 ± 0.03 | |
Table 1 Sampling information and genetic information of natural populations of Lycium ruthenicum in the Qaidam Basin
| 种群名称及编码 Population name and code | 海拔 Altitude (m) | 经纬度 Latitude and Longitude | 采样数 Sample size | 单倍型及个体数 Haplotype and the individual numbers | 单倍型多样性 Haplotype diversity (Hd ± SD) | 核苷酸多样性 Nucleotide diversity (π ± SD) | |
|---|---|---|---|---|---|---|---|
| 德令哈市红光村 Hongguang Village of Delingha City (DLH) | 2 970 | 37.38° N, 97.34° E | 10 | H1 (2), H4 (8) | 0.356 ± 0.025 | 0.000 73 ± 0.33 | |
| 都兰县诺木洪林业站 Nuomuhong Forestry Station of Dulan County (NMH1) | 2 820 | 36.41° N, 96.45° E | 15 | H5 (7), H6 (6), H7 (2) | 0.604 ± 0.069 | 0.000 46 ± 0.09 | |
| 都兰县诺木洪乡田格力村 Tiangeli Village of Nuomuhong Township of Dulan County (NMH2) | 2 762 | 36.39° N, 96.19° E | 12 | H5 (5), H7 (7) | 0.530 ± 0.053 | 0.000 37 ± 0.05 | |
| 格尔木市新乐村 Xinle Village of Golmud City (GEM1) | 2 787 | 36.39° N, 94.86° E | 12 | H1 (7), H2 (4), H3 (1) | 0.591 ± 0.011 | 0.001 26 ± 0.03 | |
| 格尔木市大格勒乡 Dagele Township of Golmud City (GEM2) | 2 837 | 36.44° N, 95.75° E | 11 | H1 (3), H2 (8) | 0.436 ± 0.018 | 0.000 09 ± 0.03 | |
| 单倍型 Haplotype | 变异位点 Variable nucleotide sites | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| psbA-trnH | psbK-psbI | trnV | ||||||||||||
| 1 | 10 | 217 | 318 | 319 | 333 | 365 | 371 | 421 | 426 | 528 | 595 | 1039 | 1208 | |
| H1 | A | C | C | G | G | T | - | G | G | G | - | T | T | G |
| H2 | ● | ● | ● | ● | ● | ● | - | ● | ● | C | A | ● | C | ● |
| H3 | ● | - | A | ● | ● | ● | - | ● | ● | ● | - | ● | C | ● |
| H4 | ● | ● | ● | ● | ● | A | C | ● | A | ● | - | ● | ● | ● |
| H5 | - | ● | ● | C | C | ● | - | C | A | ● | A | C | ● | ● |
| H6 | - | ● | ● | C | C | ● | - | C | A | ● | A | C | ● | T |
| H7 | - | ● | ● | C | C | ● | - | C | A | ● | A | C | C | ● |
Table 2 Variable nucleotide sites of seven chloroplast haplotypes of Lycium ruthenicum in the Qaidam Basin
| 单倍型 Haplotype | 变异位点 Variable nucleotide sites | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| psbA-trnH | psbK-psbI | trnV | ||||||||||||
| 1 | 10 | 217 | 318 | 319 | 333 | 365 | 371 | 421 | 426 | 528 | 595 | 1039 | 1208 | |
| H1 | A | C | C | G | G | T | - | G | G | G | - | T | T | G |
| H2 | ● | ● | ● | ● | ● | ● | - | ● | ● | C | A | ● | C | ● |
| H3 | ● | - | A | ● | ● | ● | - | ● | ● | ● | - | ● | C | ● |
| H4 | ● | ● | ● | ● | ● | A | C | ● | A | ● | - | ● | ● | ● |
| H5 | - | ● | ● | C | C | ● | - | C | A | ● | A | C | ● | ● |
| H6 | - | ● | ● | C | C | ● | - | C | A | ● | A | C | ● | T |
| H7 | - | ● | ● | C | C | ● | - | C | A | ● | A | C | C | ● |
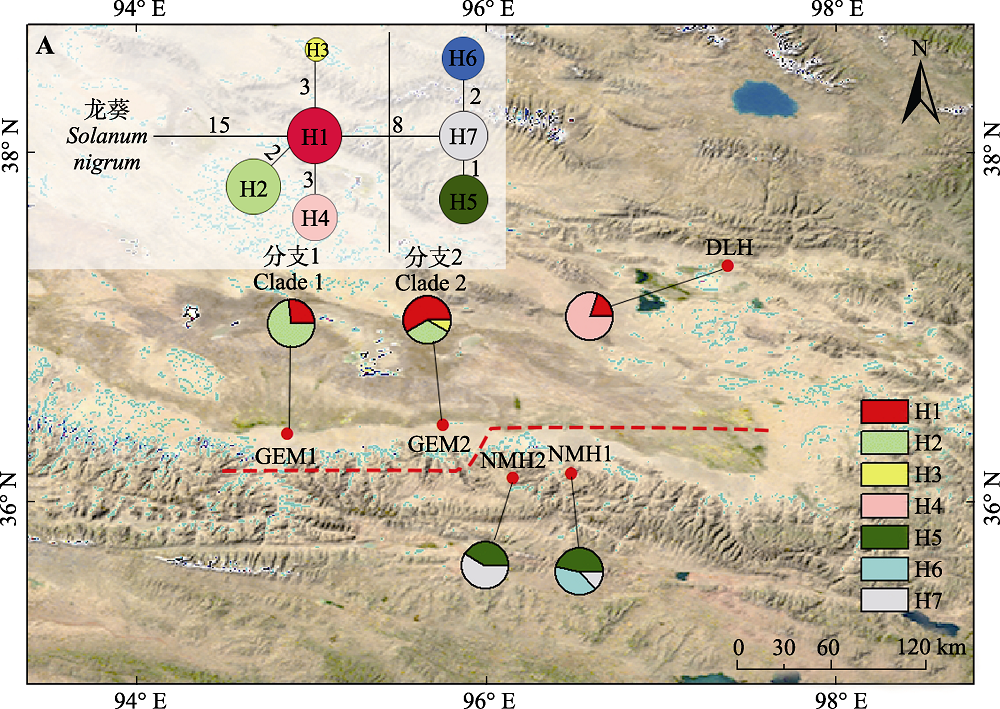
Fig. 1 Geographical distribution and the haplotype network of seven chloroplast haplotypes (H1-H7) of Lycium ruthenicum in the Qaidam Basin. The population codes in this figure are consistent with Table 1. Pie graphs indicate the frequency of each haplotype at these locations. A, In the median-joining haplotypes network, the sizes of the circles in the network are proportional to the haplotype frequencies. Branch lengths are roughly proportional to the number of mutation steps between haplotypes and nodes. The true number of steps is shown near the corresponding branch sections. Solanum nigrum was used as outgroup.
| 变异来源 Source of variation | 自由度 df | 平方和 Sum of squares | 变异组成 Variance components | 变异所占比例 Percentage of variation (%) | 固定指数 Fixation index |
|---|---|---|---|---|---|
| 种群间 Among populations | 4 | 105.305 | 2.159 37 | 80.01 | FST = 0.800 07 |
| 种群内 Within populations | 55 | 29.679 | 0.539 61 | 19.99 | |
| 总变异 Total | 59 | 135.953 | 2.698 99 | ||
| 组间 Among groups | 1 | 100.383 | 3.254 96 | 82.04 | FCT = 0.820 43 |
| 组内种群间 Among populations within groups | 3 | 6.917 | 0.145 46 | 3.67 | FSC = 0.204 18 |
| 种群内 Within populations | 56 | 28.408 | 0.566 95 | 14.29 | FST = 0.857 10 |
| 总变异 Total | 60 | 135.003 | 3.967 37 |
Table 3 Analysis of molecular variance (AMOVA) of Lycium ruthenicum in the Qaidam Basin
| 变异来源 Source of variation | 自由度 df | 平方和 Sum of squares | 变异组成 Variance components | 变异所占比例 Percentage of variation (%) | 固定指数 Fixation index |
|---|---|---|---|---|---|
| 种群间 Among populations | 4 | 105.305 | 2.159 37 | 80.01 | FST = 0.800 07 |
| 种群内 Within populations | 55 | 29.679 | 0.539 61 | 19.99 | |
| 总变异 Total | 59 | 135.953 | 2.698 99 | ||
| 组间 Among groups | 1 | 100.383 | 3.254 96 | 82.04 | FCT = 0.820 43 |
| 组内种群间 Among populations within groups | 3 | 6.917 | 0.145 46 | 3.67 | FSC = 0.204 18 |
| 种群内 Within populations | 56 | 28.408 | 0.566 95 | 14.29 | FST = 0.857 10 |
| 总变异 Total | 60 | 135.003 | 3.967 37 |
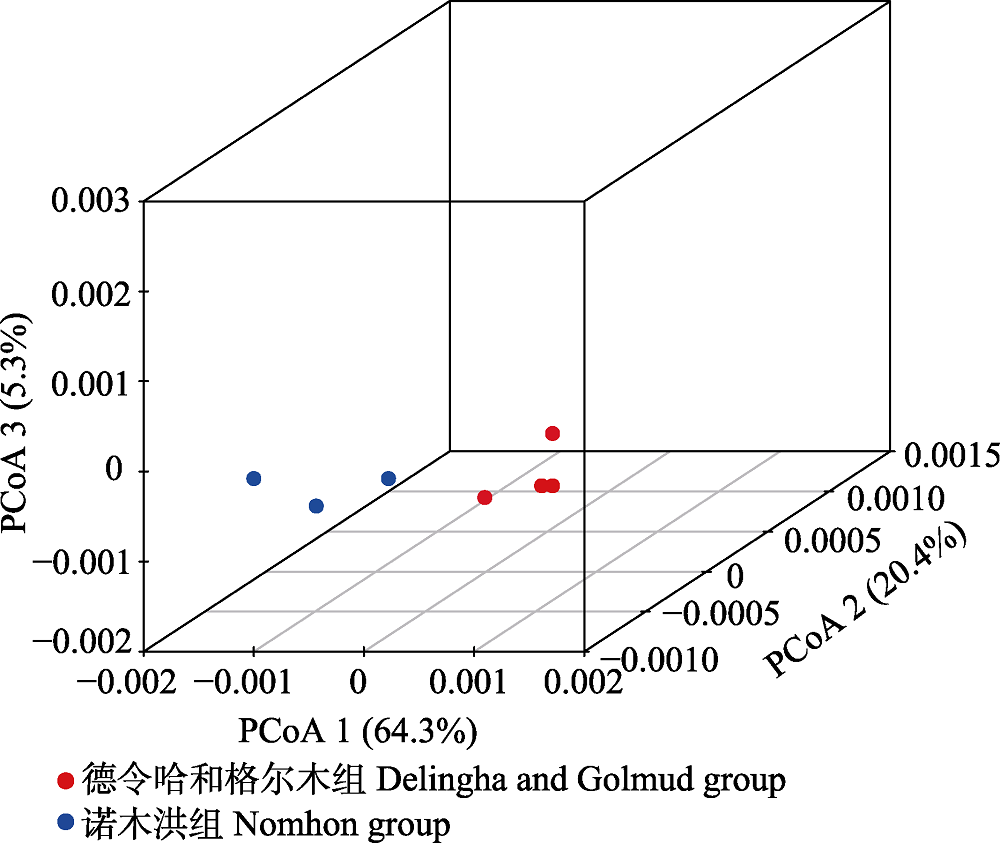
Fig. 2 Plots of the first three coordinates of the principal coordinates analysis (PCoA) at the population level for Lycium ruthenicum in the Qaidam Basin.
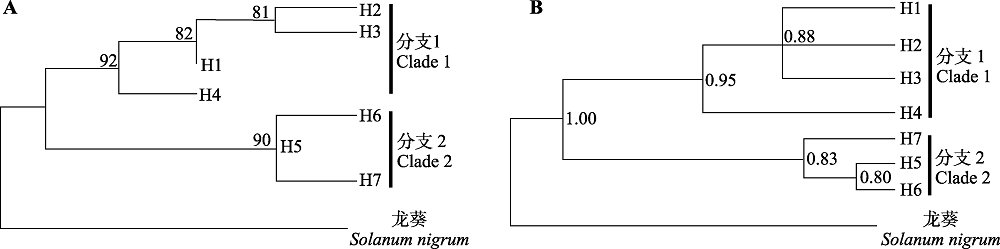
Fig. 3 Phylogenetic trees of chloroplast haplotypes of Lycium ruthenicum in the Qaidam Basin. A, Maximum likelihood (ML) tree. Bootstrap values equal to or greater than 80 are shown above the corresponding branching points. B, Bayesian tree. The values on the right of the branching points represent the posterior probability greater than 0.80. The black bars on the right of the two phylogenetic trees indicate the corresponding clades.
| [1] | Alitong Q, Jin XF, Ye ZM, Wang QF, Chen JM, Yang CF (2014). Reproductive ecology research on populations of a medicinal plant Lycium ruthenicum Murr. from Xinjiang reveals factors affecting fruit production. Plant Science Journal, 32, 570-576. |
| [ 阿力同·其米克, 金晓芳, 叶忠铭, 王青锋, 陈进明, 杨春锋 (2014). 新疆产药用植物黑果枸杞有性生殖产出差异的繁殖生态学研究. 植物科学学报, 32, 570-576.] | |
| [2] | Alitong Q, Wang QF, Yang CF, Chen JM (2013). ISSR analysis on genetic diversity of medically important Lycium ruthenicum Murr. in Xinjiang. Plant Science Journal, 31, 517-524. |
| [ 阿力同•其米克, 王青锋, 杨春锋, 陈进明 (2013). 新疆产药用植物黑果枸杞遗传多样性的ISSR分析. 植物科学学报, 31, 517-524.] | |
| [3] |
Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W (2002). Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics, 161, 1307-1320.
URL PMID |
| [4] |
Ebert D, Peakall R (2009). Chloroplast simple sequence repeats (cpSSRs): technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Molecular Ecology Resources, 9, 673-690.
DOI URL PMID |
| [5] |
Excoffier L, Lischer HEL (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567.
DOI URL PMID |
| [6] |
Flannery ML, Mitchell FJ, Coyne S, Kavanagh TA, Burke JI, Salamin N, Dowding P, Hodkinson TR (2006). Plastid genome characterisation in Brassica and Brassicaceae using a new set of nine SSRs. Theoretical and Applied Genetics, 113, 1221-1231.
DOI URL PMID |
| [7] | Fukuda T, Yokoyama J, Ohashi H (2001). Phylogeny and Biogeography of the Genus Lycium (Solanaceae): inferences from chloroplast DNA sequences. Molecular Phylogenetics & Evolution, 19, 246-258. |
| [8] | Hao YY, Xie YW, Zhang WP, Ning BS, Lu XJ (2016). The research progress on desert Lycium ruthenicum. Pratacultural Science, 33, 1835-1845. |
| [ 郝媛媛, 颉耀文, 张文培, 宁宝山, 路新军 (2016). 荒漠黑果枸杞研究进展. 草业科学, 33, 1835-1845.] | |
| [9] | He WG, Nasongcaoketu, Wu QE, Wu CH, Zhao J, Wang Y, Li YX (2015). Natural distribution and biological characteristics of Lycium ruthenicum in Yanqi Basin of Xinjiang. Chinese Wild Plant Resources, 34, 59-63. |
| [ 何文革, 那松曹克图, 吾其尔, 吴春焕, 赵洁, 王瑛, 李玉霞 (2015). 新疆焉耆盆地黑果枸杞自然分布特点及其生物特性. 中国野生植物资源, 34, 59-63.] | |
| [10] | Kuang KR, Lu AM (2011). Flora of China. Vol. 67(1). Science Press, Beijing. 8-11. |
| [ 匡可任, 路安民 (1978). 中国植物志 67卷第一分册. 科学出版社, 北京. 8-11.] | |
| [11] | Liu GY, Qi YY, Zhu CY, Chen XY, Chen JF (2016). Phenotypic diversity of wild Lycium ruthenicum populations in Qaidam Basin. Nonwood Forest Research, 34, 57-62. |
| [ 刘桂英, 祁银燕, 朱春云, 陈雪妍, 陈进福 (2016). 柴达木盆地野生黑果枸杞的表型多样性. 经济林研究, 34, 57-62.] | |
| [12] | Liu RL, Yang HW, Si JH (2011). Effect of different growth regulator on raising seedlings of Lycium ruthenicum Murr by hardwood cuttings. Journal of Anhui Agricultural Sciences, 39, 11447-11448. |
| [ 刘荣丽, 杨海文, 司剑华 (2011). 不同的生长调节剂对黑果枸杞硬枝扦插育苗的影响. 安徽农业科学, 39, 11447-11448.] | |
| [13] | Liu SW (1996). Flora of Qinhai (Solanaceae·Lycium). Qinghai People’s Press, Xining. 68. |
| [ 刘尚武 (1996). 青海植物志茄科·枸杞属. 青海人民出版社, 西宁. 68.] | |
| [14] | Liu ZG, Kang HL, Yue HL, Mei LJ, Tao YD, Shao Y (2018). Resources investigation of Lycium ruthenicum Murr. and analysis of fruits proanthocyanidin from different regions. Lishizhen Medicine and Materia Medica Research, 29, 1713-1716. |
| [ 刘增根, 康海林, 岳会兰, 梅丽娟, 陶燕铎, 邵贇 (2018). 黑果枸杞资源调查及其原花青素含量差异分析. 时珍国医国药, 29, 1713-1716.] | |
| [15] | Liu ZG, Shu QY, Wang L, Yu MF, Hu YP, Zhang HG, Tao YD, Shao Y (2012). Genetic diversity of the endangered and medically important Lycium ruthenicum Murr. revealed by sequence-related amplified polymorphism (SRAP) markers. Biochemical Systematics and Ecology, 45, 86-97. |
| [16] | Ma SM, Nie YB, Jiang XL, Xu Z, Ji WQ (2019). Genetic structure of the endangered, relict shrub Amygdalus mongolica(Rosaceae) in arid Northwest China. Australian Journal of Botany, 67, 128-139. |
| [17] |
McCauley DE (1995). The use of chloroplast DNA polymorphism in studies of gene flow in plants. Trends in Ecology & Evolution, 10, 198-202.
DOI URL PMID |
| [18] |
Miller MP (2005). Alleles in space (AIS): computer software for the joint analysis of interindividual spatial and genetic information. Journal of Heredity, 96, 722-724.
DOI URL PMID |
| [19] | Peakall R, Smouse PE (2006). Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6, 288-295. |
| [20] |
Pons O, Petit RJ (1996). Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics, 144, 1237-1245.
URL PMID |
| [21] | Qi YY, Hao GJ, Chen JF (2018) Investigation on germplasm resources of wild Lycium ruthenicum in Qinghai. Science and Technology of Qinghai Agriculture and Forestry, (3), 38-42. |
| [ 祁银燕, 郝广婧, 陈进福 (2018). 青海省野生黑果枸杞种质资源调查. 青海农林科技, (3), 38-42.] | |
| [22] |
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539-542.
DOI URL PMID |
| [23] |
Sang T, Crawford DJ, Stuessy TF (1997). Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia(Paeoniaceae). American Journal of Botany, 84, 1120-1136.
DOI URL |
| [24] | Simmons MP, Ochoterena H (2000). Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology, 2, 369-381. |
| [25] |
Shaw J, Lickey EB, Schilling EE, Small RL (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. American Journal of Botany, 94, 275-288.
DOI URL PMID |
| [26] | Sun FF, Nie YB, Ma SM, Wei B, Ji WQ (2019). Species differentiation of Haloxylon ammodendron and Haloxylon persicum based on ITS and cpDNA sequences. Scientia Silvae Sinicae, 55(3), 43-53. |
| [ 孙芳芳, 聂迎彬, 马松梅, 魏博, 吉万全 (2019). 基于ITS和cpDNA序列的梭梭和白梭梭物种分化. 林业科学, 55(3), 43-53.] | |
| [27] |
Tamura K, Dudley J, Nei M, Kumar S (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596-1599.
DOI URL PMID |
| [28] | The General Office of People’s Government of Qinghai Province (2017). Guidelines of the general office of the people’s government of Qinghai Province on strengthening the management of Lycium ruthenicum resources. Qinghai Prataculture, 26(1), 70-71. |
| [ 青海省人民政府办公厅 (2017). 青海省人民政府办公厅关于加强野生黑果枸杞资源管理工作的指导意见. 青海草业, 26(1), 70-71.] | |
| [29] | Wang JN, Chen JF, Chen WS, Zhou XY, Xu D, Li JH, Qi X (2015). Population genetic diversity of wild Lycium ruthenicum in Qaidam inferred from AFLP markers. Chinese Journal of Plant Ecology, 39, 1003-1011. |
| [ 王锦楠, 陈进福, 陈武生, 周新洋, 许东, 李际红, 亓晓 (2015). 柴达木地区野生黑果枸杞种群遗传多样性的AFLP分析. 植物生态学报, 39, 1003-1011.] | |
| [30] | Xin JP, Zhu CY (2015). Morphological structure comparison of Lycium ruthenium at different salt habitats in Qaidam Basin. Journal of West China Forestry Science, 44, 73-78. |
| [ 辛菊平, 朱春云 (2015). 柴达木盆地不同盐生境下黑果枸杞形态结构比较. 西部林业科学, 44, 73-78.] |
| [1] | DONG Shao-Qiong, HOU Dong-Jie, QU Xiao-Yun, GUO Ke. A plot-based dataset of plant communities on the Qaidam Basin, China [J]. Chin J Plant Ecol, 2024, 48(4): 534-540. |
| [2] | YAN Han, ZHANG Yun-Ling, MA Song-Mei, WANG Chun-Cheng, ZHANG Dan. Suitable distribution simulation and local environmental adaptability differentiation of Lycium ruthenicum in Xinjiang, China [J]. Chin J Plant Ecol, 2021, 45(11): 1221-1230. |
| [3] | OU Wen-Hui, LIU Ya-Heng, LI Na, XU Zhi-Yan, PENG Qiu-Tong, YANG Yu-Jing, LI Zhong-Qiang. Testing multiple hypotheses for the richness pattern of macrophyte in the Qaidam Basin of Northwest China [J]. Chin J Plant Ecol, 2021, 45(11): 1213-1220. |
| [4] | JI Ruo-Xuan, YU Xiao, CHANG Yuan, SHEN Chao, BAI Xue-Qia, XIA Xin-Li, YIN Wei-Lun, LIU Chao. Geographical provenance variation of leaf anatomical structure of Caryopteris mongholica and its significance in response to environmental changes [J]. Chin J Plant Ecol, 2020, 44(3): 277-286. |
| [5] | WANG Jin-Nan,CHEN Jin-Fu,Chen Wu-Sheng,Zhou Xin-Yang,XU Dong,LI Ji-Hong,QI Xiao. Population genetic diversity of wild Lycium ruthenicum in Qaidam inferred from AFLP markers [J]. Chin J Plan Ecolo, 2015, 39(10): 1003-1011. |
| [6] | Sun Shi-zhou. The Vegetation of Qaidam Basin and its Surrounding Mountains [J]. Chin J Plan Ecolo, 1989, 13(3): 236-249. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Chinese Journal of Plant Ecology
Tel: 010-62836134, 62836138, E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn