

Chin J Plant Ecol ›› 2007, Vol. 31 ›› Issue (4): 561-567.DOI: 10.17521/cjpe.2007.0072
Special Issue: 青藏高原植物生态学:种群生态学; 生物多样性
• Articles • Previous Articles Next Articles
LU Jian-Ying1, MA Rui-Jun2,*( ), SUN Kun1
), SUN Kun1
Received:2006-07-06
Accepted:2006-12-11
Online:2007-07-06
Published:2007-07-30
Contact:
MA Rui-Jun
LU Jian-Ying, MA Rui-Jun, SUN Kun. CLONAL DIVERSITY AND STRUCTURE IN POLYGONUM VIVIPARUM[J]. Chin J Plant Ecol, 2007, 31(4): 561-567.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.plant-ecology.com/EN/10.17521/cjpe.2007.0072
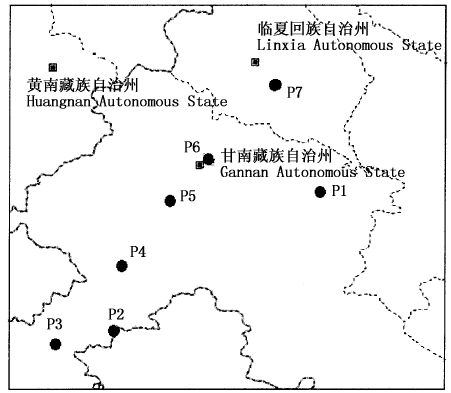
Fig.1 The distribution map of study population of Polygonum viviparum Showing sampling places of seven populations of Polygonum viviparum P1: 冶力关-临潭 Yeliguan-Lintan P2: 尕玛梁 Gamaliang P3: 玛曲 Maqu P4: 尕海-碌曲 Gahai-Luqu P5: 碌曲-合作 Luqu-Hezuo P6: 依毛梁 Yimaoliang P7: 和政南阳山 Hezheng Nanyangshan Mountain
| 种群 Population | 海拔 Altitude (m) | 采样个数 No. of sample | 生境 Habitats |
|---|---|---|---|
| P1 | 2 500 | 25 | 山上 On the Mountain |
| P2 | 3 900 | 25 | 路边草地 Roodside meadow |
| P3 | 3 400 | 25 | 草地 Meadow |
| P4 | 3 150 | 20 | 草地 Meadow |
| P5 | 3 000 | 27 | 湿地 March |
| P6 | 2 900 | 24 | 湿地 March |
| P7 | 2 000 | 25 | 河滩 Beach |
Table 1 The habitats in sampling sites of Polygonum viviparum
| 种群 Population | 海拔 Altitude (m) | 采样个数 No. of sample | 生境 Habitats |
|---|---|---|---|
| P1 | 2 500 | 25 | 山上 On the Mountain |
| P2 | 3 900 | 25 | 路边草地 Roodside meadow |
| P3 | 3 400 | 25 | 草地 Meadow |
| P4 | 3 150 | 20 | 草地 Meadow |
| P5 | 3 000 | 27 | 湿地 March |
| P6 | 2 900 | 24 | 湿地 March |
| P7 | 2 000 | 25 | 河滩 Beach |
| 引物 Primer | 序列(5'-3') Sequence (5'-3') | DNA总位点数 Total number of loci | 多态位点数 No. of polymorphic loci |
|---|---|---|---|
| S5 | TGCGCCCTTC | 10 | 7 |
| S8 | GTCCACACGG | 13 | 7 |
| S12 | CCTTGACGCA | 7 | 6 |
| S13 | TTCCCCCGCT | 8 | 6 |
| S223 | CTCCCTGCAA | 7 | 4 |
| S393 | ACCGCCTGCT | 8 | 5 |
| S494 | GGACGCTTCA | 10 | 6 |
| S500 | TCGCCCAGTC | 11 | 7 |
| S1102 | ACTTGACGGG | 10 | 8 |
| S1107 | AACCGCGGCA | 7 | 6 |
| S1156 | CACAACGGGA | 10 | 9 |
| S1420 | CTTCTCGGAC | 8 | 7 |
| S2113 | CCGCCGGTAA | 8 | 6 |
Table 2 The sequence of primers and the number of tested loci
| 引物 Primer | 序列(5'-3') Sequence (5'-3') | DNA总位点数 Total number of loci | 多态位点数 No. of polymorphic loci |
|---|---|---|---|
| S5 | TGCGCCCTTC | 10 | 7 |
| S8 | GTCCACACGG | 13 | 7 |
| S12 | CCTTGACGCA | 7 | 6 |
| S13 | TTCCCCCGCT | 8 | 6 |
| S223 | CTCCCTGCAA | 7 | 4 |
| S393 | ACCGCCTGCT | 8 | 5 |
| S494 | GGACGCTTCA | 10 | 6 |
| S500 | TCGCCCAGTC | 11 | 7 |
| S1102 | ACTTGACGGG | 10 | 8 |
| S1107 | AACCGCGGCA | 7 | 6 |
| S1156 | CACAACGGGA | 10 | 9 |
| S1420 | CTTCTCGGAC | 8 | 7 |
| S2113 | CCGCCGGTAA | 8 | 6 |
| 种群 Population | 样本大小(N) Sample size | 种群基株(G) Number of genets | 平均克隆大(N/G) Average size of genotype | 基因型比率(PD) Proportion of distinct genotypes | Simpson多样性指数(D) Simpson diversity index | 均匀度(E) Fager's evenness |
|---|---|---|---|---|---|---|
| P1 | 20 | 10 | 2.000 | 0.500 | 0.879 | 0.712 |
| P2 | 20 | 8 | 2.500 | 0.400 | 0.816 | 0.684 |
| P3 | 20 | 4 | 5.000 | 0.200 | 0.500 | 0.427 |
| P4 | 20 | 5 | 4.000 | 0.250 | 0.800 | 0.911 |
| P5 | 20 | 7 | 2.857 | 0.350 | 0.689 | 0.441 |
| P6 | 20 | 4 | 5.000 | 0.200 | 0.284 | 0.000 |
| P7 | 20 | 5 | 4.000 | 0.250 | 0.505 | 0.288 |
| 平均Mean | 20 | 6 | 3.622 | 0.307 | 0.639 | 0.495 |
Table 3 Clonal diversity in all populations
| 种群 Population | 样本大小(N) Sample size | 种群基株(G) Number of genets | 平均克隆大(N/G) Average size of genotype | 基因型比率(PD) Proportion of distinct genotypes | Simpson多样性指数(D) Simpson diversity index | 均匀度(E) Fager's evenness |
|---|---|---|---|---|---|---|
| P1 | 20 | 10 | 2.000 | 0.500 | 0.879 | 0.712 |
| P2 | 20 | 8 | 2.500 | 0.400 | 0.816 | 0.684 |
| P3 | 20 | 4 | 5.000 | 0.200 | 0.500 | 0.427 |
| P4 | 20 | 5 | 4.000 | 0.250 | 0.800 | 0.911 |
| P5 | 20 | 7 | 2.857 | 0.350 | 0.689 | 0.441 |
| P6 | 20 | 4 | 5.000 | 0.200 | 0.284 | 0.000 |
| P7 | 20 | 5 | 4.000 | 0.250 | 0.505 | 0.288 |
| 平均Mean | 20 | 6 | 3.622 | 0.307 | 0.639 | 0.495 |
| 种群 Population | 克隆数 Number of clones | 构型 Clonal architecture | 种群内克隆分布情况(不同克隆的植株用‘/’隔开) Clones distribution within population (different clones distingwished by `/') |
|---|---|---|---|
| P1 | 10 | 过渡型 Transition | 1,2/3,4,5,6,8,10/7/9/11,13/12/14/15-17,19/18/20 |
| P2 | 8 | 过渡型 Transition | 1,3,5,7,8,9,12,18/2,4,6/10/11/13/14/15-17/19,20 |
| P3 | 4 | 密集型 Phalanx | 1-11,15,16,17/12,14/13/18-20 |
| P4 | 5 | 游击型 Guerilla | 1,3,6,11,12,13,16/2,4,5,14,17/7,15/8-10/18-20 |
| P5 | 7 | 游击型 Guerilla | 1,2/3,4,6,7,9,10,12,15,16,18,20/5/8/11,14,19/13/17 |
| P6 | 4 | 密集型 Phalanx | 1-4,6-15,17,19,20/5/16/18 |
| P7 | 5 | 密集型 Phalanx | 1/2-4,6-15,17/5/16/18-20 |
Table 4 Clones number and clonal architecture within populations
| 种群 Population | 克隆数 Number of clones | 构型 Clonal architecture | 种群内克隆分布情况(不同克隆的植株用‘/’隔开) Clones distribution within population (different clones distingwished by `/') |
|---|---|---|---|
| P1 | 10 | 过渡型 Transition | 1,2/3,4,5,6,8,10/7/9/11,13/12/14/15-17,19/18/20 |
| P2 | 8 | 过渡型 Transition | 1,3,5,7,8,9,12,18/2,4,6/10/11/13/14/15-17/19,20 |
| P3 | 4 | 密集型 Phalanx | 1-11,15,16,17/12,14/13/18-20 |
| P4 | 5 | 游击型 Guerilla | 1,3,6,11,12,13,16/2,4,5,14,17/7,15/8-10/18-20 |
| P5 | 7 | 游击型 Guerilla | 1,2/3,4,6,7,9,10,12,15,16,18,20/5/8/11,14,19/13/17 |
| P6 | 4 | 密集型 Phalanx | 1-4,6-15,17,19,20/5/16/18 |
| P7 | 5 | 密集型 Phalanx | 1/2-4,6-15,17/5/16/18-20 |
| [1] | Barret SCH, Shore JS (1989). Isozyme variation in colonizing plants. In: Soltis DE, Soltis PS eds. Isozymes in Plant Biology, Chapman & Hall, Lando, 106-126. |
| [2] |
Bauert MR (1993). Vivipary in Polygonum viviparum: an adaption to cold climate? Nordic Journal of Botany, 13, 473-480.
DOI URL |
| [3] | Chen JH (陈锦华), Wang XF (汪小凡), Lü YT (吕应堂) (2003). Pattern of clonal growth in a natural population of Sagittaria pygmaea Miq. (Alismataceae). ( Journal of Wuhan University (Natural Science Edition) (武汉大学学报(理学版)), 49, 523-527. (in Chinese with English abstract) |
| [4] | Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry Bulletin, 9, 11-15. |
| [5] |
Eckert CG, Barrett SCH (1993). Clonal reproduction and pattern of genotypic diversity in Decodon verticillatus (Lythraceae). American Journal of Botany, 80, 1175-1182.
DOI URL |
| [6] |
Ellstrand NC, Roose ML (1987). Patterns of genotypic diversity in clonal plant species. American Journal of Botany, 74, 123-131.
DOI URL |
| [7] |
Fager EW (1972). Diversity: a sampling study. The American Nature, 106, 293-310.
DOI URL |
| [8] |
GilI DE, Lin C, Perkins SL (1995). Genetic mosaicism in plants and animals. Annual Review of Ecology and Systematics, 26, 423-444.
DOI URL |
| [9] | Gray A (1993). The vascular plant pioneers of primary successions: persistence and phenotypic plasticity, In: Miles J, Walton D eds. Primary Succession on Land. Blackwell, Oxford, UK, 179-191. |
| [10] | Hangelbroek HH, Ouborg NJ, Santamaría L, Schwenk K (2002). Clonal diversity and structure within a population of the pondweed Potamogeton pectinatus foraged by Bewick's swans. Molecular Ecology, 12, 2137-2150. |
| [11] | Hartmann H (1957). Studien über die vegetative Fortpflanzung in den Hochalpen. Jahresbericht der Naturforschenden Gesellschaft Graubündens, 86, 3-168. |
| [12] | Klimes L, Klimesova J, Hendriks R, van Groenendael J, (1997). Clonal plant architecture: a comparative analysis of form and function. In: de Kroon H, van Groenendael J eds. The Ecology and Evolution of Clonal Plants. Backhuys Publishers, Leiden, the Netherlands, 1-29. |
| [13] | Li YZ (李有忠), Shen SD (沈颂东) (1996). A preliminary morphological and anatomical investigation in the vegetative and reproductive organs of Polygonum viviparum. Journal of Qinghai Normal University (Natural Science Edition) (青海师范大学学报(自然科学版)), 1, 34-40. (in Chinese) |
| [14] |
Lynch M, Milligan BG (1994). Analysis of population genetic structure with RAPD markers. Molecular Ecology, 3, 91-99.
DOI URL PMID |
| [15] |
Montalvo AM, Conard SG, Conkle MT, Hodgskiss PD (1997). Population structure, genetic diversity, and clone formation in Quercus chrysolepis (Fagaceae). American Journal of Botany, 84, 1553-1564.
URL PMID |
| [16] | Nei M (1973). Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the United States of American, 70, 3321-3323. |
| [17] |
Parker KC, Hamrick JL (1992). Genetic diversity and clonal structure in a columnar cactus, Lophocereus schottii. American Journal of Botany, 79, 86-96.
DOI URL |
| [18] | Pielou EC (1969). An Introduction to Mathematical Ecology. Wiley-Interscience, New York. |
| [19] | Ruan CJ (阮成江), He ZX (何祯祥), Zhou CF (周长芳) (2005). Plant Molecular Ecology (植物分子生态学). Chemical Industry Press, Beijing. (in Chinese) |
| [20] |
Sipes SD, Wolf PG (1997). Clonal structure and patterns of allozyme diversity in the rare endemic Cycladenia humilis var. jonessii (Apocynaceae). American Journal of Botany, 84, 401-409.
URL PMID |
| [21] |
Stenstrom A, Jonsson BO, Jonsdottir IS, Fagerstrom T, Augner M (2001). Genetic variation and clonal diversity in four clonal sedges (Carex) along the Arctic coast of Eurasia. Molecular Ecology, 10, 497-513.
DOI URL PMID |
| [22] |
Støcklin J, B⁉umler E (1996). Seed rain, seedling establishment, and clonal growth strategies on a glacier foreland. Journal of Vegetation Science, 7, 45-56.
DOI URL |
| [23] |
Suyama Y, Obayashi K, Hayashi I (2000). Clonal structure in a dwarf bamboo (Sasasenanesis) population inferred from amplified fragment length polymorphism (AFLP) fingerprints. Molecular Ecology, 9, 901-906.
DOI URL PMID |
| [24] |
Torimaru T, Tomaru N, Nishimura N, Yamamoto S (2003). Clonal diversity and genetic differentiation in Ilex leucoclada M. patches in an old-growth beech forest. Molecular Ecology, 12, 809-818.
DOI URL PMID |
| [25] |
Younga AG, Hill JH, Murray BG, Peakall R (2002). Breeding system, genetic diversity and clonal structure in the subalpine forb Rutidosis leiolepis F. Muell. (Asteraceae). Biological Conservation, 106, 71-78.
DOI URL |
| [26] | Zhang FM (张富民), Ge S (葛颂) (2002). Data analysis in population genetics.Ⅰ. Analysis of RAPD data with AMOVA. Biodiversity Science (生物多样性), 10, 438-444. (in Chinese with English abstract) |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Chinese Journal of Plant Ecology
Tel: 010-62836134, 62836138, E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn