

植物生态学报 ›› 2007, Vol. 31 ›› Issue (4): 680-688.DOI: 10.17521/cjpe.2007.0088
收稿日期:2006-08-20
接受日期:2007-01-05
出版日期:2007-08-20
发布日期:2007-07-30
通讯作者:
董鸣
作者简介:*E-mail: dongming@ibcas.ac.cn基金资助:
LI Jun-Min1,3, DONG Ming2,*( ), ZHONG Zhang-Cheng1
), ZHONG Zhang-Cheng1
Received:2006-08-20
Accepted:2007-01-05
Online:2007-08-20
Published:2007-07-30
Contact:
DONG Ming
摘要:
利用简单重复序列区间(Inter simple sequence repeat, ISSR)分子标记技术分析了入侵植物薇甘菊(Mikania micrantha)8个种群的遗传多样性及遗传分化。12个引物共扩增出171个位点,其中多态位点有103个,多态位点百分率(P%)为60.23%,Shannon信息指数(I)为0.281 8,Nei指数(h)为0.184 9,薇甘菊在物种水平具有较高的遗传多样性。AMOVA显示薇甘菊具有较高的遗传分化,36.49%的变异发生在种群间,63.51%的变异发生于种群内,基因分化系数(GST)为0.352 4。种群间的基因流较高,为0.918 7。薇甘菊8个种群之间的遗传相似性很高,平均为0.915 5;遗传距离很小,平均为0.088 4。采用UPGMA法对8个种群进行聚类,可以将8个种群分为两大类群,即内伶仃岛为一个类群,而深圳与东莞内陆种群组成另一类群。薇甘菊现有遗传结构的形成与其生活史特性及入侵生态学特性有关。
李钧敏, 董鸣, 钟章成. 入侵植物薇甘菊种群的遗传分化. 植物生态学报, 2007, 31(4): 680-688. DOI: 10.17521/cjpe.2007.0088
LI Jun-Min, DONG Ming, ZHONG Zhang-Cheng. POPULATION GENETIC DIFFERENTIATIONS IN THE INVASIVE PLANT MIKANIA MICRANTHA IN CHINA. Chinese Journal of Plant Ecology, 2007, 31(4): 680-688. DOI: 10.17521/cjpe.2007.0088
| 引物 Primers | 序列 Sequences | 引物 Primers | 序列 Sequences |
|---|---|---|---|
| UBC808 | (AG)8C | UBC873 | (GACA)4 |
| UBC811 | (GA)8C | UBC880 | (GGAGA)3 |
| UBC841 | (GA)8YC | UBC884 | HBH(AG)7 |
| UBC857 | (AC)8YG | UBC885 | BHB(GA)7 |
| UBC864 | (ATG)6 | UBC886 | VDV(CT)7 |
| UBC868 | (GAA)6 | UBC887 | DVD(TC)7 |
表1 ISSR分析用的12个引物序列
Table 1 Sequences of 12 primers used in ISSR analysis
| 引物 Primers | 序列 Sequences | 引物 Primers | 序列 Sequences |
|---|---|---|---|
| UBC808 | (AG)8C | UBC873 | (GACA)4 |
| UBC811 | (GA)8C | UBC880 | (GGAGA)3 |
| UBC841 | (GA)8YC | UBC884 | HBH(AG)7 |
| UBC857 | (AC)8YG | UBC885 | BHB(GA)7 |
| UBC864 | (ATG)6 | UBC886 | VDV(CT)7 |
| UBC868 | (GAA)6 | UBC887 | DVD(TC)7 |
图1 薇甘菊去梧桐山公路边(GL)种群引物UBC 811扩增结果
Fig.1 ISSR amplification of Mikania micrantha in GL population generated with primer UBC 811 1~20: 个体1~20 Individual 1-20 M: λDNA/EcoR I+Hind III分子量参照物 λDNA/EcoR I+Hind III standard molecular weight marker
| 种群(代码) Population(Code) | 多态位点数 Number of polymorphic loci | 多态位点百分率(%) Percetage of polymorphic loci (P) | Shannon信息指数 Shannon's information index (I) | Nei基因多样性 Nei's genet diversity (h) |
|---|---|---|---|---|
| 西湖乐园 (XH) | 69 | 40.35 | 0.233 5 | 0.159 8 |
| 民俗村 (MS) | 59 | 34.50 | 0.179 1 | 0.118 9 |
| 大王山 (DWS) | 62 | 36.26 | 0.191 2 | 0.129 5 |
| 大望村 (DW) | 58 | 33.92 | 0.190 1 | 0.128 4 |
| 内伶仃岛I (NLD I) | 55 | 32.16 | 0.155 2 | 0.102 6 |
| 内伶仃岛II (NLD II) | 57 | 33.33 | 0.165 8 | 0.110 8 |
| 梧桐山 (WT) | 58 | 33.92 | 0.164 0 | 0.108 2 |
| 去梧桐山公路边 (GL) | 55 | 32.16 | 0.152 2 | 0.099 7 |
| 种Speices | 103 | 60.23 | 0.281 8 | 0.184 9 |
表2 薇甘菊种群的遗传多样性
Table 2 Estimates of genetic diversity within populations of Mikania micrantha
| 种群(代码) Population(Code) | 多态位点数 Number of polymorphic loci | 多态位点百分率(%) Percetage of polymorphic loci (P) | Shannon信息指数 Shannon's information index (I) | Nei基因多样性 Nei's genet diversity (h) |
|---|---|---|---|---|
| 西湖乐园 (XH) | 69 | 40.35 | 0.233 5 | 0.159 8 |
| 民俗村 (MS) | 59 | 34.50 | 0.179 1 | 0.118 9 |
| 大王山 (DWS) | 62 | 36.26 | 0.191 2 | 0.129 5 |
| 大望村 (DW) | 58 | 33.92 | 0.190 1 | 0.128 4 |
| 内伶仃岛I (NLD I) | 55 | 32.16 | 0.155 2 | 0.102 6 |
| 内伶仃岛II (NLD II) | 57 | 33.33 | 0.165 8 | 0.110 8 |
| 梧桐山 (WT) | 58 | 33.92 | 0.164 0 | 0.108 2 |
| 去梧桐山公路边 (GL) | 55 | 32.16 | 0.152 2 | 0.099 7 |
| 种Speices | 103 | 60.23 | 0.281 8 | 0.184 9 |
| 变异来源 Source of variance | 自由度 df | 变异组分 Variance component | 百分率 Pcentage | p值 p value* |
|---|---|---|---|---|
| 种群间Among popualtions | 7 | 5.257 7 | 36.49 | <0.001 |
| 种群内Within populations | 152 | 9.150 7 | 63.51 | <0.001 |
表3 8个薇甘菊种群的AMOVA分析
Table 3 Analysis of molecular variance (AMOVA) for the eight populations of Mikania micrantha
| 变异来源 Source of variance | 自由度 df | 变异组分 Variance component | 百分率 Pcentage | p值 p value* |
|---|---|---|---|---|
| 种群间Among popualtions | 7 | 5.257 7 | 36.49 | <0.001 |
| 种群内Within populations | 152 | 9.150 7 | 63.51 | <0.001 |
| Shannon信息指数Shannon's information index | Nei基因多样性 Nei's gene diversity | ||
|---|---|---|---|
| 种群内遗传多样性 Within population genetic diversity, Hpop(SD) | 0.184 9 (0.037 8) | 种群内基因多样性 Within population gene diversity, HS (SD) | 0.119 7 (0.017 4) |
| 种群总的遗传多样性 Total genetic diversity, Hsp(SD) | 0.281 8 (0.275 3) | 种群总的基因多样性 Total gene diversity, HT (SD) | 0.184 9 (0.194 3) |
| 种群内遗传多样性比率 Ratio of genetic diversity within population, Hpop/Hsp | 0.656 1 | 种群内基因多样性比率 Ratio of gene diversity within population, Hs/HT | 0.647 6 |
| 种群间遗传多样性比率 Ratio of genetic diversity among population, (Hsp-Hpop)/Hsp | 0.343 9 | 遗传分化系数 Gene differentiation among populations, GST | 0.3524 |
| 基因流 Gene flow, Nm | 0.918 7 |
表4 8个薇甘菊种群的遗传分化
Table 4 Genetic differentiations within and among the eight populations of Mikania micrantha
| Shannon信息指数Shannon's information index | Nei基因多样性 Nei's gene diversity | ||
|---|---|---|---|
| 种群内遗传多样性 Within population genetic diversity, Hpop(SD) | 0.184 9 (0.037 8) | 种群内基因多样性 Within population gene diversity, HS (SD) | 0.119 7 (0.017 4) |
| 种群总的遗传多样性 Total genetic diversity, Hsp(SD) | 0.281 8 (0.275 3) | 种群总的基因多样性 Total gene diversity, HT (SD) | 0.184 9 (0.194 3) |
| 种群内遗传多样性比率 Ratio of genetic diversity within population, Hpop/Hsp | 0.656 1 | 种群内基因多样性比率 Ratio of gene diversity within population, Hs/HT | 0.647 6 |
| 种群间遗传多样性比率 Ratio of genetic diversity among population, (Hsp-Hpop)/Hsp | 0.343 9 | 遗传分化系数 Gene differentiation among populations, GST | 0.3524 |
| 基因流 Gene flow, Nm | 0.918 7 |
| 种群Population | XH | MS | DWS | DW | NLD I | NLD II | WT | GL |
|---|---|---|---|---|---|---|---|---|
| 西湖乐园(XH) | - | 0.912 2 | 0.927 9 | 0.916 8 | 0.913 9 | 0.904 6 | 0.928 5 | 0.913 3 |
| 民俗村(MS) | 0.091 9 | - | 0.900 2 | 0.920 4 | 0.902 5 | 0.894 2 | 0.925 7 | 0.900 3 |
| 大王山(DWS) | 0.074 8 | 0.105 1 | - | 0.924 7 | 0.906 2 | 0.904 8 | 0.928 5 | 0.895 4 |
| 大望村(DW) | 0.086 9 | 0.083 0 | 0.078 3 | - | 0.922 8 | 0.911 2 | 0.933 9 | 0.915 3 |
| 内伶仃岛I (NLD I) | 0.090 1 | 0.102 6 | 0.098 5 | 0.080 4 | - | 0.943 1 | 0.920 9 | 0.893 |
| 内伶仃岛II (NLD II) | 0.100 3 | 0.111 8 | 0.100 0 | 0.093 0 | 0.058 5 | - | 0.930 9 | 0.893 5 |
| 梧桐山(WT) | 0.074 2 | 0.077 2 | 0.074 2 | 0.068 4 | 0.082 4 | 0.071 6 | - | 0.950 0 |
| 去梧桐山公路边(GL) | 0.090 6 | 0.105 0 | 0.110 4 | 0.088 5 | 0.113 2 | 0.112 6 | 0.051 3 | - |
表5 8个薇甘菊种群间的遗传相似度与遗传距离
Table 5 Genetic similarity and genetic distances among the eight populations of Mikania micrantha
| 种群Population | XH | MS | DWS | DW | NLD I | NLD II | WT | GL |
|---|---|---|---|---|---|---|---|---|
| 西湖乐园(XH) | - | 0.912 2 | 0.927 9 | 0.916 8 | 0.913 9 | 0.904 6 | 0.928 5 | 0.913 3 |
| 民俗村(MS) | 0.091 9 | - | 0.900 2 | 0.920 4 | 0.902 5 | 0.894 2 | 0.925 7 | 0.900 3 |
| 大王山(DWS) | 0.074 8 | 0.105 1 | - | 0.924 7 | 0.906 2 | 0.904 8 | 0.928 5 | 0.895 4 |
| 大望村(DW) | 0.086 9 | 0.083 0 | 0.078 3 | - | 0.922 8 | 0.911 2 | 0.933 9 | 0.915 3 |
| 内伶仃岛I (NLD I) | 0.090 1 | 0.102 6 | 0.098 5 | 0.080 4 | - | 0.943 1 | 0.920 9 | 0.893 |
| 内伶仃岛II (NLD II) | 0.100 3 | 0.111 8 | 0.100 0 | 0.093 0 | 0.058 5 | - | 0.930 9 | 0.893 5 |
| 梧桐山(WT) | 0.074 2 | 0.077 2 | 0.074 2 | 0.068 4 | 0.082 4 | 0.071 6 | - | 0.950 0 |
| 去梧桐山公路边(GL) | 0.090 6 | 0.105 0 | 0.110 4 | 0.088 5 | 0.113 2 | 0.112 6 | 0.051 3 | - |
| 种群Population | XH | MS | DWS | DW | NLD I | NLD II | WT | GL |
|---|---|---|---|---|---|---|---|---|
| 西湖乐园(XH) | - | |||||||
| 民俗村(MS) | 50.3 | - | ||||||
| 大王山(DWS) | 26 | 26.3 | - | |||||
| 大望村(DW) | 54.7 | 19 | 37.6 | - | ||||
| 内伶仃岛I (NLD I) | 103.9 | 22.1 | 29.1 | 40.5 | - | |||
| 内伶仃岛II (NLD II) | 104.2 | 22.5 | 29.6 | 40.7 | 1.6 | - | ||
| 梧桐山(WT) | 54.2 | 19.5 | 37.2 | 0.6 | 41.1 | 41.3 | - | |
| 去梧桐山公路边(GL) | 54.4 | 19.2 | 37.4 | 0.3 | 40.9 | 41.1 | 0.3 | - |
表6 8个薇甘菊种群间的地理距离(km)
Table 6 Geographical distances (km) among the eight populations of Mikania micrantha
| 种群Population | XH | MS | DWS | DW | NLD I | NLD II | WT | GL |
|---|---|---|---|---|---|---|---|---|
| 西湖乐园(XH) | - | |||||||
| 民俗村(MS) | 50.3 | - | ||||||
| 大王山(DWS) | 26 | 26.3 | - | |||||
| 大望村(DW) | 54.7 | 19 | 37.6 | - | ||||
| 内伶仃岛I (NLD I) | 103.9 | 22.1 | 29.1 | 40.5 | - | |||
| 内伶仃岛II (NLD II) | 104.2 | 22.5 | 29.6 | 40.7 | 1.6 | - | ||
| 梧桐山(WT) | 54.2 | 19.5 | 37.2 | 0.6 | 41.1 | 41.3 | - | |
| 去梧桐山公路边(GL) | 54.4 | 19.2 | 37.4 | 0.3 | 40.9 | 41.1 | 0.3 | - |
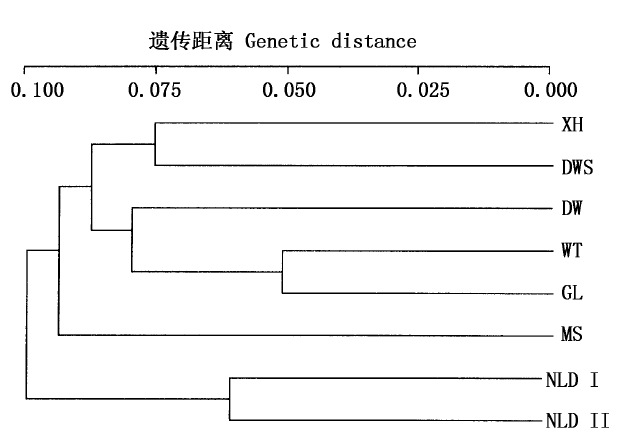
图2 8个薇甘菊种群间的遗传距离聚类图
Fig.2 Dengrogram produced by POPGENE software for the eight populations of Mikania micrantha XH、MS、DWS、DW、NLD I、NLD II、WT、GL同表2 See Table 2
| [1] |
Aguinagalde I, Hampe A, Mohanty A, Martín JP, Duminil J, Petit RJ (2005). Effects of life-history traits and species distribution on genetic structure at maternally inherited markers in European trees and shrubs. Journal of Biogeography, 32, 329-339.
DOI URL |
| [2] | Ammiraju JSS, Dholakin BB, Sntra DK, Singh H, Lagu MD, Tamhankar SA, Dhaiual HS, Rao VS, Gupta VS, Ranjekar PK (2001). Identification of inter simple sequence repeat (ISSR)markers associated with seed size in wheat. Theoretical Application of Genetics, 102, 726-732. |
| [3] | Bakir M, Facey PC, Hassan I, Mulder WH, Porter RB (2004). Mikanolide from Jamaican Mikania micrantha. Acta Crystallographica, Section C, Crystal Structure Communications, 60, 798-800. |
| [4] | Brown AHD, Marshall DR (1981). Evolutionary changes accompanying colonization in plants. In: Scudder GCE, Reveal JL eds. Colonization, Succession and Stability. Blackwell Scientific Publications, Oxford, |
| [5] |
Esselman EJ, Jianqiang L, Crawford DJ, Windus JL, Wolfe AD (1999). Clonal diversity in the rare Calamagrostis porteri spp. insperata (Poaceae): comparative results for allozymes and random amplified polymorphic DNA (RAPD) and intersimple sequence repeat (ISSR) markers. Molecular Ecology, 8, 443-451.
DOI URL |
| [6] |
Excoffier L, Smouse PF, Quattro JM (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131, 479-491.
URL PMID |
| [7] | Feng HL( 冯惠玲), Cao HL(曹洪麟), Liang XD(梁晓东), Zhou X(周霞), Ye WH(叶万辉) (2002). The distribution and harmful effect of Mikania micrantha in Guangdong. Journal of Tropical and Subtropical Botany (热带亚热带植物学报), 10, 263-270. (in Chinese with English abstract) |
| [8] |
Godt MJW, Hamrick JL(1991). Genetic variation in Lathyrus latifolius (Leguminosae). American Journal of Botany, 78, 1163-1171.
DOI URL |
| [9] |
Govindaraju DR (1988). Relationship between dispersal ability and levels of gene flow in plants. Oikos, 52, 31-35.
DOI URL |
| [10] | Hamrick JL, Godt MJW (1989). Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS eds. Plant Population Genetics, Breeding, and Genetic Resources. Sinauer, Sunderland, USA, |
| [11] | Hamrick JL, Godt MJW (1990). Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS eds. Plant Population Genetics, Breeding, and Genetic Resources. Sinauer, Sunderland, USA. |
| [12] |
Hamrick JL, Godt MJW, Sherman-Broyles SL (1992). Factors influencing levels of genetic diversity in woody plant species. New Forests, 6, 95-124.
DOI URL |
| [13] |
Hamrick JL, Linhart YB, Mitton JB (1979). Relationships between life history characteristics and electrophoretically detectable genetic variation in plants. Annual Review of Ecological Systematics, 10, 173-200.
DOI URL |
| [14] |
Hollingsworth ML, Hollingsworth PM, Jenkins GI, Bailey JP, Ferris C (1998). The use of molecular markers to study patterns of genotypic diversity in some invasive alien Fallopia spp. (Polygonaceae). Molecular Ecology, 7, 1681-1691.
DOI URL |
| [15] | Hu YJ (胡玉佳), Paul PHB (毕培曦) (1994). A study on life cycle and response to herbicides of Mikania micrantha. Acta Scientiarum Naturalium Universitatis Sunyatseni (中山大学学报(自然科学版)) , 33(4) , 88-95. (in Chinese with English abstract) |
| [16] |
Ismail BS, Chong TV (2002). Effects of aqueous extracts and decomposition of Mikania micrantha H.B.K. debris on selected agronomic crops. Weed Biology and Management, 2, 31-38.
DOI URL |
| [17] | Kong GH(孔国辉), Wu QG(吴七根), Hu QM(胡启明), Ye WH(叶万辉) (2000a). Further supplementary data on Mikania micrantha H.B.K. (Asteraceae). Journal of Tropical and Subtropical Botany (热带亚热带植物学报), 8(2), 128-130. (in Chinese with English abstract) |
| [18] | Kong GH(孔国辉), Wu QG(吴七根), Hu QM(胡启明) (2000b). Emergence of invasive weed Mikania micrantha H.B.K. in our county. Journal of Tropical and Subtropical Botany (热带亚热带植物学报), 8(1), 27. (in Chinese) |
| [19] | Li JM(李钧敏), Ke SS(柯世省), Jin ZX(金则新) (2002). Extraction and determination of DNA from Heptacodium miconioides. Guihaia (广西植物), 22, 499-502. (in Chinese with English abstract) |
| [20] | Li MG(李鸣光), Zhang WY(张炜银), Wang BS(王伯荪), Zhang JL(张军丽), Zan QJ(昝启杰), Wang YJ(王勇军) (2002). A preliminary study on the seed germination in Mikania micrantha. Acta Scientiarum Naturalium Universitis Sunyatseni (中山大学学报(自然科学版)), 41(6), 57-59. (in Chinese with English abstract) |
| [21] |
Li WG, Shen JJ, Wang JB (2005). Genetic diversity of the annual weed Monochoria vaginalis in southern China detected by random amplified polymorphic DNA and inter-simple sequence repeat analyses. Weed Research, 45, 424-430.
DOI URL |
| [22] | Li A, Ge S (2001). Genetic variation and colonal diversity of Psammochloa villosa (Poaceae) detected by ISSR markers. Annual of Botany, 87, 585-590. |
| [23] | Ma CL(马翠兰), Liu XS(刘星释), Zhang YL(张玉兰) (2001). Application of DNA molecular markers to fruit trees. Journal of Inner Mongolia Agriculature University (Natural Science Edition) (内蒙古农业大学报(自然科学版)), 22(2), 105-112. (in Chinese with English abstract) |
| [24] |
Meekins JF, Ballard HE, McCarthy BC (2001). Genetic variation and molecular biogeography of a North American invasive plant species ( Alliaria Petiolata, Brassicaceae). International Journal of Plant Science, 162, 161-169.
DOI URL |
| [25] |
Mooney HA, Cleland EE (2001). The evolutionary impact of invasive species. Proceedings of the National Academy Sciences of the United States of America, 98, 5446-5451.
DOI URL |
| [26] |
Nei M (1972). Genetic distance between populations. American Naturalist, 106, 283-292.
DOI URL |
| [27] |
Nei M, Maruyama T, Chakraborty R (1975). The bottleneck effect and genetic variability in populations. Evolution, 29, 1-10.
DOI URL PMID |
| [28] |
Nicollier G, Thompson AC (1981). Essential oil and terpenoids of Mikania micrantha. Phytochemistry, 20, 2587-2588.
DOI URL |
| [29] |
Nissen SJ, Masters RA, Lee DJ, Rowe ML(1995). DNA-based marker systems to determine genetic diversity of weedy species and their application to biocontrol. Weed Science, 43, 504-513.
DOI URL |
| [30] |
Novak SJ, Mack RN (1993). Genetic variation in Bromus tectorum (Poaceae): comparison between native and introduced populations. Heredity, 71, 167-176.
DOI URL |
| [31] |
Pappert RA, Hamrick JL, Donovan LA (2000). Genetic variation in Pueraria lobata (Fabaceae), an introduced, clonal, invasive plant of the southeastern United States. American Journal of Botany, 87, 1240-1245.
URL PMID |
| [32] |
Ren MX, Zhang QG, Zhang DY (2005). Random amplified polymorphic DNA markers reveal low genetic variation and a single dominant genotype in Eichhornia crassipes populations throughout China. Weed Research, 45, 236-244.
DOI URL |
| [33] |
Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, Cabin RJ, Cohen JE, Ellstrand NC, McCauley DE, O'Neil P, Parker IM, Thompson JN, Weller SG (2001). The population biology of invasive species. Annual Review of Ecology and Systematics, 32, 305-332.
DOI URL |
| [34] |
Schierenbeck KA, Hamrick JL, Mack RN (1995). Comparison of allozyme variability in a native and an introduced species of Lonicera. Heredity, 75, 1-9.
DOI URL |
| [35] | Shao H(邵华), Peng SL(彭少麟), Liu YX(刘运笑), Zhang C(张弛), Xiang YC(向言词) (2002). The biological control and the natural enemy of Mikania micrantha H.B.K.'s in China. Ecologic Science (生态科学), 21, 33-36. (in Chinese with English abstract) |
| [36] | Shao WT(邵婉婷), Han SC(韩诗畴), Huang SS(黄寿山), Liu WH(刘文惠), Li KH(李开煌), Peng TX(彭统序), Li LY(李丽英) (2002). Progress on the control of invasive weed Mikania micrantha. Guangdong Agricultural Sciences (广东农业科学), 37(1), 43-45. (in Chinese) |
| [37] | Swamy PS, Ramakrishnan PS (1987a). Weed potential of Mikania micrantha H.B.K., and its control in fallows after shifting agriculture (Jhum) in North-East India. Agriculture, Ecosystems and Environment, 18, 195-204. |
| [38] |
Swamy PS, Ramakrishnan PS (1987b). Contribution of Mikania micrantha during secondary succession following slash-and-burn agriculture (Jhum) in North-East India. I. Biomass litterfall and productivity. Forest Ecology and Management, 22, 229-237.
DOI URL |
| [39] | Tsumura Y, Ohba K, Strauss SH (1996). Diversity and inheritance of inter-simple sequence repeat polymorphisms in Douglas fir (Pseudotsuga menzesii) and sugi (Cryptomeria japonica). Theoretical Application of Genetics, 92, 40-45. |
| [40] | Wain RP, Haller WT, Martin DF (1984). Genetic relationship among two forms of alligator weed. Journal of Aquatic Plant Management, 22, 104-105. |
| [41] | Wang BS(王伯荪), Liao WB(廖文波), Miao RH(缪汝槐) (2001). Revision of Mikania from China and the key of four relative species. Acta Scientiarum Naturalium Universitis Sunyatseni (中山大学学报(自然科学版)), 40(5), 72-75. (in Chinese with English abstract) |
| [42] | Wang BS(王伯荪), Wang YJ(王勇军), Liao WB(廖文波) (2004). Invasion Ecology and Control on Mikania micrantha (外来杂草薇甘菊的入侵生态及其治理). Science Press, Beijing. (in Chinese) |
| [43] |
Wei XY, Huang HJ, Wu P, Cao HL, Ye WH (2004). Phenolic constituents from Mikania micrantha. Biochemical Systematics and Ecology, 32, 1091-1096.
DOI URL |
| [44] | Wen DZ (温达志), Ye WH (叶万辉), Feng HL (冯惠玲), Cai CX (蔡楚雄) (2000). Comparison of basic photosynthetic characteristics between exotic invader weed Mikania micrantha and its companion species. Journal of Tropical and Subtropical Botany (热带亚热带植物学报) , 8, 139-146. (in Chinese with English abstract) |
| [45] |
Wolfe AD, Xiang QY, Kephart SR (1998). Assessing hybridization in natural populations of Penstemon (Scrophulariaceae) using hypervariable intersimple sequence repeat (ISSR) bands. Molecular Ecology, 7, 1107-1125.
DOI URL PMID |
| [46] | Wright S 1978. Evolution and the Genetics of Populations Vol 4. Variability Within and Among Natural Populations. University of Chicago Press, Chicago. |
| [47] | Yang QH(杨期和), Feng HL(冯惠玲), Ye WH (叶万辉), Cao HL(曹洪麟), Deng X(邓雄), Xu KY(许凯扬) (2003). An investigation of the effects of environmental factors on the flowering and seed setting of Mikania micrantha H.B.K. (Compositae). Journal of Tropical and Subtropical Botany (热带亚热带植物学报), 11, 123-126. (in Chinese with English abstract) |
| [48] |
Ye WH, Li J, Cao HL, Ge XJ (2003). Genetic uniformity of Alternanthera philoxeroides in South China. Weed Research, 43, 297-302.
DOI URL |
| [49] |
Ye WH, Mu HP, Cao HL, Ge XJ (2004). Genetic structure of the invasive Chromolaena odorata in China. Weed Research, 44, 129-135.
DOI URL |
| [50] | Yeh FC, Boyle TJB (1997). Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belgian Journal of Botany, 129, 157. |
| [51] | Zan QJ(昝启杰), Wang BS(王伯荪), Wang YJ(王勇军), Liao WB(廖文波), Li MG(李鸣光), Xu HL(徐华林) (2002). The ecological evaluation on the controlling Mikania micrantha by Cuscuta campestris. Acta Scientiarum Natralium Universitis Sunyatseni (中山大学学报(自然科学版)), 41(6), 60-63. (in Chinese with English abstract) |
| [52] | Zan QJ(昝启杰), Wang YJ(王勇军), Wang BS (王伯荪), Liao WB (廖文波), Li MG (李鸣光) (2000). The distribution and harm of the exotic weed Mikania micarantha. Chinese Journal of Ecology (生态学杂志), 19(6), 58-61. (in Chinese with English abstract) |
| [53] | Zhang FM(张富民), Ge S(葛颂) (2002). Data analysis in population genetics. I. Analysis of RAPD data with AMOVA. Biodiversity Science (生物多样性), 10, 438-444. (in Chinese with English abstract) |
| [54] | Zhang WY(张炜银), Wang BS(王伯荪), Li MG(李鸣光), Zan QJ(昝启杰), Wang YJ(王勇军) (2002a). The effects of light intensity on growth and morphology in Mikania micrntha seedlings. Sun Yatsen University Forum (中山大学学报论丛), 22(1), 222-226. (in Chinese with English abstract) |
| [55] | Zhang WY(张炜银), Wang BS(王伯荪), Liao WB(廖文波), Li MG(李鸣光), Wang YJ(王勇军), Zan QJ(昝启杰) (2002b). Progress in studies on an exotic vicious weed Mikania micrantha. Chinese Journal of Applied Ecology (应用生态学报), 13, 1684-1688. (in Chinese with English abstract) |
| [56] | Zhang WY(张炜银), Wang BS(王伯荪), Zhang JL(张军丽), Li MG(李鸣光), Zan QJ(昝启杰), Wang YJ(王勇军) (2002c). Study on the structure and dynamics of seedlings of Mikania micrantha populations. Acta Scientiarum Naturalium Universitis Sunyatseni (中山大学学报(自然科学版)), 41(6), 64-66. (in Chinese with English abstract) |
| [57] |
Zietkiewicz E, Rafalski A, Labuda D (1994). Genome fingerprinting by simple sequence repeat (SSR)-anhorcd polymerase chain reaction amplification. Genomics, 20, 176-183.
DOI URL PMID |
| [1] | 汪晶晶, 王嘉浩, 黄致云, Vanessa Chiamaka OKECHUKW, 胡蝶, 祁珊珊, 戴志聪, 杜道林. 不同氮水平下内生固氮菌对入侵植物南美蟛蜞菊生长策略的影响[J]. 植物生态学报, 2023, 47(2): 195-205. |
| [2] | 陈天翌, 娄安如. 青藏高原东侧白桦种群的遗传多样性与遗传结构[J]. 植物生态学报, 2022, 46(5): 561-568. |
| [3] | 杜军, 王文, 何志斌, 陈龙飞, 蔺鹏飞, 朱喜, 田全彦. 祁连山青海云杉物候表型的空间分异及其内在机制[J]. 植物生态学报, 2021, 45(8): 834-843. |
| [4] | 秦文超, 陶至彬, 王永健, 刘艳杰, 黄伟. 资源脉冲对外来植物入侵影响的研究进展和展望[J]. 植物生态学报, 2021, 45(6): 573-582. |
| [5] | 林秦文, 于胜祥, 唐赛春, 崔夏, 高信芬, 王焕冲, 刘全儒, 马金双. 中国外来归化植物的编目现状及有关问题[J]. 植物生态学报, 2021, 45(11): 1275-1280. |
| [6] | 庞芳, 夏维康, 何敏, 祁珊珊, 戴志聪, 杜道林. 固氮菌缓解氮限制环境中丛枝菌根真菌对加拿大一枝黄花的营养竞争[J]. 植物生态学报, 2020, 44(7): 782-790. |
| [7] | 唐金琦, 郭小城, 鲁新瑜, 刘明超, 张海艳, 冯玉龙, 孔德良. 外来入侵植物对本地植物菌根真菌的影响及其机制[J]. 植物生态学报, 2020, 44(11): 1095-1112. |
| [8] | 闫雅楠, 叶小齐, 吴明, 闫明, 张昕丽. 入侵植物加拿大一枝黄花根际解钾菌多样性及解钾活性[J]. 植物生态学报, 2019, 43(6): 543-556. |
| [9] | 张新新, 王茜, 胡颖, 周玮, 陈晓阳, 胡新生. 植物边缘种群遗传多样性研究进展[J]. 植物生态学报, 2019, 43(5): 383-395. |
| [10] | 张俪文, 韩广轩. 植物遗传多样性与生态系统功能关系的研究进展[J]. 植物生态学报, 2018, 42(10): 977-989. |
| [11] | 古春凤, 叶小齐, 吴明, 邵学新, 焦盛武. 草甘膦对加拿大一枝黄花和伴生植物白茅种间竞争关系的影响[J]. 植物生态学报, 2017, 41(4): 439-449. |
| [12] | 杨雪, 申俊芳, 赵念席, 高玉葆. 不同基因型羊草数量性状的可塑性及遗传分化[J]. 植物生态学报, 2017, 41(3): 359-368. |
| [13] | 彭扬, 彭培好, 李景吉. 模拟氮沉降对矢车菊属植物Centaurea stoebe种群生长和竞争能力的影响[J]. 植物生态学报, 2016, 40(7): 679-685. |
| [14] | 罗文启, 符少怀, 杨小波, 陈玉凯, 周威, 杨琦, 陶楚, 周文嵩. 海南岛入侵植物的分布特点及其对本地植物的影响[J]. 植物生态学报, 2015, 39(5): 486-500. |
| [15] | 王锦楠, 陈进福, 陈武生, 周新洋, 许东, 李际红, 亓晓. 柴达木地区野生黑果枸杞种群遗传多样性的AFLP分析[J]. 植物生态学报, 2015, 39(10): 1003-1011. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
Copyright © 2022 版权所有 《植物生态学报》编辑部
地址: 北京香山南辛村20号, 邮编: 100093
Tel.: 010-62836134, 62836138; Fax: 010-82599431; E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
备案号: 京ICP备16067583号-19