

植物生态学报 ›› 2007, Vol. 31 ›› Issue (4): 561-567.DOI: 10.17521/cjpe.2007.0072
所属专题: 青藏高原植物生态学:种群生态学; 生物多样性
收稿日期:2006-07-06
接受日期:2006-12-11
出版日期:2007-07-06
发布日期:2007-07-30
通讯作者:
马瑞君
作者简介:*E-mail:ruijunma2003@yahoo.com.cn基金资助:
LU Jian-Ying1, MA Rui-Jun2,*( ), SUN Kun1
), SUN Kun1
Received:2006-07-06
Accepted:2006-12-11
Online:2007-07-06
Published:2007-07-30
Contact:
MA Rui-Jun
摘要:
珠芽蓼(Polygonum viviparum)是青藏高原东缘广泛分布的克隆植物,具有有性和无性(根状茎和珠芽)两种生殖方式。该研究采用RAPD技术对分布于不同海拔的珠芽蓼7个自然种群进行了克隆结构和克隆多样性(是单克隆种群还是多克隆种群)以及克隆多样性与海拔因子之间的相关性研究,为了解高山克隆植物对环境的适应性策略及揭示克隆植物的繁殖和分布特点提供科学依据。研究结果表明:1)采用13条RAPD引物对珠芽蓼7个种群共140个样本进行扩增分析,共扩增到117个位点,其中多态性位点84个,多态位点百分率PPL达到71.79%,检测到43个基因型,且全部为局限型基因型;2)与Ellstrand和Roose(1987)总结的克隆植物的克隆多样性平均值相比(PD=0.17,D=0.62),珠芽蓼种群克隆多样性水平稍高,Simpson指数平均为0.639,基因型比率PD平均为0.307;3)克隆结构分析表明,珠芽蓼种群内克隆之间的镶嵌明显,这可能与珠芽蓼过渡型的克隆构型有关。研究中珠芽蓼种群的构型有游击型、密集型以及这两者之间的过渡类型;4)采用SPSS软件对珠芽蓼种群的克隆多样性与海拔高度进行相关性分析,结果显示它们之间并无明显的相关性。
陆建英, 马瑞君, 孙坤. 珠芽蓼种群克隆多样性及克隆结构的初步研究. 植物生态学报, 2007, 31(4): 561-567. DOI: 10.17521/cjpe.2007.0072
LU Jian-Ying, MA Rui-Jun, SUN Kun. CLONAL DIVERSITY AND STRUCTURE IN POLYGONUM VIVIPARUM. Chinese Journal of Plant Ecology, 2007, 31(4): 561-567. DOI: 10.17521/cjpe.2007.0072
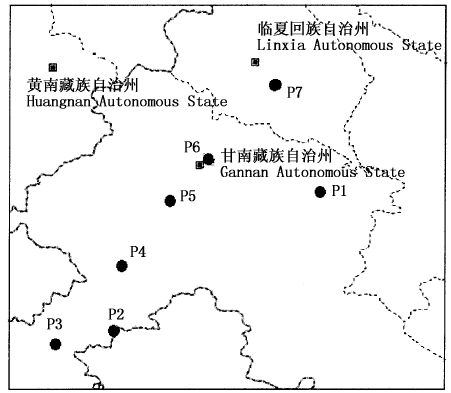
图1 珠芽蓼种群采样分布图 ●表示珠芽蓼7个种群的采样地点
Fig.1 The distribution map of study population of Polygonum viviparum Showing sampling places of seven populations of Polygonum viviparum P1: 冶力关-临潭 Yeliguan-Lintan P2: 尕玛梁 Gamaliang P3: 玛曲 Maqu P4: 尕海-碌曲 Gahai-Luqu P5: 碌曲-合作 Luqu-Hezuo P6: 依毛梁 Yimaoliang P7: 和政南阳山 Hezheng Nanyangshan Mountain
| 种群 Population | 海拔 Altitude (m) | 采样个数 No. of sample | 生境 Habitats |
|---|---|---|---|
| P1 | 2 500 | 25 | 山上 On the Mountain |
| P2 | 3 900 | 25 | 路边草地 Roodside meadow |
| P3 | 3 400 | 25 | 草地 Meadow |
| P4 | 3 150 | 20 | 草地 Meadow |
| P5 | 3 000 | 27 | 湿地 March |
| P6 | 2 900 | 24 | 湿地 March |
| P7 | 2 000 | 25 | 河滩 Beach |
表1 珠芽蓼天然种群RAPD分析的样品及其生境条件
Table 1 The habitats in sampling sites of Polygonum viviparum
| 种群 Population | 海拔 Altitude (m) | 采样个数 No. of sample | 生境 Habitats |
|---|---|---|---|
| P1 | 2 500 | 25 | 山上 On the Mountain |
| P2 | 3 900 | 25 | 路边草地 Roodside meadow |
| P3 | 3 400 | 25 | 草地 Meadow |
| P4 | 3 150 | 20 | 草地 Meadow |
| P5 | 3 000 | 27 | 湿地 March |
| P6 | 2 900 | 24 | 湿地 March |
| P7 | 2 000 | 25 | 河滩 Beach |
| 引物 Primer | 序列(5'-3') Sequence (5'-3') | DNA总位点数 Total number of loci | 多态位点数 No. of polymorphic loci |
|---|---|---|---|
| S5 | TGCGCCCTTC | 10 | 7 |
| S8 | GTCCACACGG | 13 | 7 |
| S12 | CCTTGACGCA | 7 | 6 |
| S13 | TTCCCCCGCT | 8 | 6 |
| S223 | CTCCCTGCAA | 7 | 4 |
| S393 | ACCGCCTGCT | 8 | 5 |
| S494 | GGACGCTTCA | 10 | 6 |
| S500 | TCGCCCAGTC | 11 | 7 |
| S1102 | ACTTGACGGG | 10 | 8 |
| S1107 | AACCGCGGCA | 7 | 6 |
| S1156 | CACAACGGGA | 10 | 9 |
| S1420 | CTTCTCGGAC | 8 | 7 |
| S2113 | CCGCCGGTAA | 8 | 6 |
表2 用于珠芽蓼扩增的随机引物的序列及其检测的位点数
Table 2 The sequence of primers and the number of tested loci
| 引物 Primer | 序列(5'-3') Sequence (5'-3') | DNA总位点数 Total number of loci | 多态位点数 No. of polymorphic loci |
|---|---|---|---|
| S5 | TGCGCCCTTC | 10 | 7 |
| S8 | GTCCACACGG | 13 | 7 |
| S12 | CCTTGACGCA | 7 | 6 |
| S13 | TTCCCCCGCT | 8 | 6 |
| S223 | CTCCCTGCAA | 7 | 4 |
| S393 | ACCGCCTGCT | 8 | 5 |
| S494 | GGACGCTTCA | 10 | 6 |
| S500 | TCGCCCAGTC | 11 | 7 |
| S1102 | ACTTGACGGG | 10 | 8 |
| S1107 | AACCGCGGCA | 7 | 6 |
| S1156 | CACAACGGGA | 10 | 9 |
| S1420 | CTTCTCGGAC | 8 | 7 |
| S2113 | CCGCCGGTAA | 8 | 6 |
图2 引物S12扩增的P4种群的DNA片段图谱 1~20:P4种群的20个样品 20 samples of P4 population M:分子量标准(100 bp DNA ladder plus) Standard marker
Fig.2 The PCR amplification pattern of P4 population generated with primer S12
| 种群 Population | 样本大小(N) Sample size | 种群基株(G) Number of genets | 平均克隆大(N/G) Average size of genotype | 基因型比率(PD) Proportion of distinct genotypes | Simpson多样性指数(D) Simpson diversity index | 均匀度(E) Fager's evenness |
|---|---|---|---|---|---|---|
| P1 | 20 | 10 | 2.000 | 0.500 | 0.879 | 0.712 |
| P2 | 20 | 8 | 2.500 | 0.400 | 0.816 | 0.684 |
| P3 | 20 | 4 | 5.000 | 0.200 | 0.500 | 0.427 |
| P4 | 20 | 5 | 4.000 | 0.250 | 0.800 | 0.911 |
| P5 | 20 | 7 | 2.857 | 0.350 | 0.689 | 0.441 |
| P6 | 20 | 4 | 5.000 | 0.200 | 0.284 | 0.000 |
| P7 | 20 | 5 | 4.000 | 0.250 | 0.505 | 0.288 |
| 平均Mean | 20 | 6 | 3.622 | 0.307 | 0.639 | 0.495 |
表3 珠芽蓼种群的克隆多样性
Table 3 Clonal diversity in all populations
| 种群 Population | 样本大小(N) Sample size | 种群基株(G) Number of genets | 平均克隆大(N/G) Average size of genotype | 基因型比率(PD) Proportion of distinct genotypes | Simpson多样性指数(D) Simpson diversity index | 均匀度(E) Fager's evenness |
|---|---|---|---|---|---|---|
| P1 | 20 | 10 | 2.000 | 0.500 | 0.879 | 0.712 |
| P2 | 20 | 8 | 2.500 | 0.400 | 0.816 | 0.684 |
| P3 | 20 | 4 | 5.000 | 0.200 | 0.500 | 0.427 |
| P4 | 20 | 5 | 4.000 | 0.250 | 0.800 | 0.911 |
| P5 | 20 | 7 | 2.857 | 0.350 | 0.689 | 0.441 |
| P6 | 20 | 4 | 5.000 | 0.200 | 0.284 | 0.000 |
| P7 | 20 | 5 | 4.000 | 0.250 | 0.505 | 0.288 |
| 平均Mean | 20 | 6 | 3.622 | 0.307 | 0.639 | 0.495 |
| 种群 Population | 克隆数 Number of clones | 构型 Clonal architecture | 种群内克隆分布情况(不同克隆的植株用‘/’隔开) Clones distribution within population (different clones distingwished by `/') |
|---|---|---|---|
| P1 | 10 | 过渡型 Transition | 1,2/3,4,5,6,8,10/7/9/11,13/12/14/15-17,19/18/20 |
| P2 | 8 | 过渡型 Transition | 1,3,5,7,8,9,12,18/2,4,6/10/11/13/14/15-17/19,20 |
| P3 | 4 | 密集型 Phalanx | 1-11,15,16,17/12,14/13/18-20 |
| P4 | 5 | 游击型 Guerilla | 1,3,6,11,12,13,16/2,4,5,14,17/7,15/8-10/18-20 |
| P5 | 7 | 游击型 Guerilla | 1,2/3,4,6,7,9,10,12,15,16,18,20/5/8/11,14,19/13/17 |
| P6 | 4 | 密集型 Phalanx | 1-4,6-15,17,19,20/5/16/18 |
| P7 | 5 | 密集型 Phalanx | 1/2-4,6-15,17/5/16/18-20 |
表4 种群内克隆数目和构型分布
Table 4 Clones number and clonal architecture within populations
| 种群 Population | 克隆数 Number of clones | 构型 Clonal architecture | 种群内克隆分布情况(不同克隆的植株用‘/’隔开) Clones distribution within population (different clones distingwished by `/') |
|---|---|---|---|
| P1 | 10 | 过渡型 Transition | 1,2/3,4,5,6,8,10/7/9/11,13/12/14/15-17,19/18/20 |
| P2 | 8 | 过渡型 Transition | 1,3,5,7,8,9,12,18/2,4,6/10/11/13/14/15-17/19,20 |
| P3 | 4 | 密集型 Phalanx | 1-11,15,16,17/12,14/13/18-20 |
| P4 | 5 | 游击型 Guerilla | 1,3,6,11,12,13,16/2,4,5,14,17/7,15/8-10/18-20 |
| P5 | 7 | 游击型 Guerilla | 1,2/3,4,6,7,9,10,12,15,16,18,20/5/8/11,14,19/13/17 |
| P6 | 4 | 密集型 Phalanx | 1-4,6-15,17,19,20/5/16/18 |
| P7 | 5 | 密集型 Phalanx | 1/2-4,6-15,17/5/16/18-20 |
| [1] | Barret SCH, Shore JS (1989). Isozyme variation in colonizing plants. In: Soltis DE, Soltis PS eds. Isozymes in Plant Biology, Chapman & Hall, Lando, 106-126. |
| [2] |
Bauert MR (1993). Vivipary in Polygonum viviparum: an adaption to cold climate? Nordic Journal of Botany, 13, 473-480.
DOI URL |
| [3] | Chen JH (陈锦华), Wang XF (汪小凡), Lü YT (吕应堂) (2003). Pattern of clonal growth in a natural population of Sagittaria pygmaea Miq. (Alismataceae). ( Journal of Wuhan University (Natural Science Edition) (武汉大学学报(理学版)), 49, 523-527. (in Chinese with English abstract) |
| [4] | Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry Bulletin, 9, 11-15. |
| [5] |
Eckert CG, Barrett SCH (1993). Clonal reproduction and pattern of genotypic diversity in Decodon verticillatus (Lythraceae). American Journal of Botany, 80, 1175-1182.
DOI URL |
| [6] |
Ellstrand NC, Roose ML (1987). Patterns of genotypic diversity in clonal plant species. American Journal of Botany, 74, 123-131.
DOI URL |
| [7] |
Fager EW (1972). Diversity: a sampling study. The American Nature, 106, 293-310.
DOI URL |
| [8] |
GilI DE, Lin C, Perkins SL (1995). Genetic mosaicism in plants and animals. Annual Review of Ecology and Systematics, 26, 423-444.
DOI URL |
| [9] | Gray A (1993). The vascular plant pioneers of primary successions: persistence and phenotypic plasticity, In: Miles J, Walton D eds. Primary Succession on Land. Blackwell, Oxford, UK, 179-191. |
| [10] | Hangelbroek HH, Ouborg NJ, Santamaría L, Schwenk K (2002). Clonal diversity and structure within a population of the pondweed Potamogeton pectinatus foraged by Bewick's swans. Molecular Ecology, 12, 2137-2150. |
| [11] | Hartmann H (1957). Studien über die vegetative Fortpflanzung in den Hochalpen. Jahresbericht der Naturforschenden Gesellschaft Graubündens, 86, 3-168. |
| [12] | Klimes L, Klimesova J, Hendriks R, van Groenendael J, (1997). Clonal plant architecture: a comparative analysis of form and function. In: de Kroon H, van Groenendael J eds. The Ecology and Evolution of Clonal Plants. Backhuys Publishers, Leiden, the Netherlands, 1-29. |
| [13] | Li YZ (李有忠), Shen SD (沈颂东) (1996). A preliminary morphological and anatomical investigation in the vegetative and reproductive organs of Polygonum viviparum. Journal of Qinghai Normal University (Natural Science Edition) (青海师范大学学报(自然科学版)), 1, 34-40. (in Chinese) |
| [14] |
Lynch M, Milligan BG (1994). Analysis of population genetic structure with RAPD markers. Molecular Ecology, 3, 91-99.
DOI URL PMID |
| [15] |
Montalvo AM, Conard SG, Conkle MT, Hodgskiss PD (1997). Population structure, genetic diversity, and clone formation in Quercus chrysolepis (Fagaceae). American Journal of Botany, 84, 1553-1564.
URL PMID |
| [16] | Nei M (1973). Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the United States of American, 70, 3321-3323. |
| [17] |
Parker KC, Hamrick JL (1992). Genetic diversity and clonal structure in a columnar cactus, Lophocereus schottii. American Journal of Botany, 79, 86-96.
DOI URL |
| [18] | Pielou EC (1969). An Introduction to Mathematical Ecology. Wiley-Interscience, New York. |
| [19] | Ruan CJ (阮成江), He ZX (何祯祥), Zhou CF (周长芳) (2005). Plant Molecular Ecology (植物分子生态学). Chemical Industry Press, Beijing. (in Chinese) |
| [20] |
Sipes SD, Wolf PG (1997). Clonal structure and patterns of allozyme diversity in the rare endemic Cycladenia humilis var. jonessii (Apocynaceae). American Journal of Botany, 84, 401-409.
URL PMID |
| [21] |
Stenstrom A, Jonsson BO, Jonsdottir IS, Fagerstrom T, Augner M (2001). Genetic variation and clonal diversity in four clonal sedges (Carex) along the Arctic coast of Eurasia. Molecular Ecology, 10, 497-513.
DOI URL PMID |
| [22] |
Støcklin J, B⁉umler E (1996). Seed rain, seedling establishment, and clonal growth strategies on a glacier foreland. Journal of Vegetation Science, 7, 45-56.
DOI URL |
| [23] |
Suyama Y, Obayashi K, Hayashi I (2000). Clonal structure in a dwarf bamboo (Sasasenanesis) population inferred from amplified fragment length polymorphism (AFLP) fingerprints. Molecular Ecology, 9, 901-906.
DOI URL PMID |
| [24] |
Torimaru T, Tomaru N, Nishimura N, Yamamoto S (2003). Clonal diversity and genetic differentiation in Ilex leucoclada M. patches in an old-growth beech forest. Molecular Ecology, 12, 809-818.
DOI URL PMID |
| [25] |
Younga AG, Hill JH, Murray BG, Peakall R (2002). Breeding system, genetic diversity and clonal structure in the subalpine forb Rutidosis leiolepis F. Muell. (Asteraceae). Biological Conservation, 106, 71-78.
DOI URL |
| [26] | Zhang FM (张富民), Ge S (葛颂) (2002). Data analysis in population genetics.Ⅰ. Analysis of RAPD data with AMOVA. Biodiversity Science (生物多样性), 10, 438-444. (in Chinese with English abstract) |
| [1] | 杨丽婷, 谢燕燕, 左珂怡, 徐森, 谷瑞, 陈双林, 郭子武. 分株比例对异质光环境下美丽箬竹克隆系统光合生理的影响[J]. 植物生态学报, 2022, 46(1): 88-101. |
| [2] | 叶学华, 薛建国, 谢秀芳, 黄振英. 外部干扰对根茎型克隆植物甘草自然种群植株生长及主要药用成分含量的影响[J]. 植物生态学报, 2020, 44(9): 951-961. |
| [3] | 王意锟, 金爱武, 朱强根, 邱永华, 季新良, 张四海. 施肥对毛竹种群不同年龄分株间胸径大小关系的影响[J]. 植物生态学报, 2014, 38(3): 289-297. |
| [4] | 彭一可, 罗芳丽, 李红丽, 于飞海. 根状茎型植物扁秆荆三棱对土壤养分异质性尺度和对比度的生长响应[J]. 植物生态学报, 2013, 37(4): 335-343. |
| [5] | 罗维成, 曾凡江, 刘波, 宋聪, 彭守兰, Stefan K. ARNDT. 疏叶骆驼刺母株与子株间的水分整合[J]. 植物生态学报, 2013, 37(2): 164-172. |
| [6] | 葛俊, 邢福. 克隆植物对种间竞争的适应策略[J]. 植物生态学报, 2012, 36(6): 587-596. |
| [7] | 朱志红, 席博, 李英年, 臧岳铭, 王文娟, 刘建秀, 郭华. 高寒草甸不同生境粗喙薹草补偿生长研究[J]. 植物生态学报, 2010, 34(3): 348-358. |
| [8] | 张道远, 王建成, 施翔. 根茎型木本克隆植物准噶尔无叶豆的种群数量动态[J]. 植物生态学报, 2009, 33(5): 893-900. |
| [9] | 周会平, 陈进, 张寿洲. 具混合繁殖策略的草本植物异果舞花姜的居群遗传结构[J]. 植物生态学报, 2008, 32(4): 751-759. |
| [10] | 王琼, 廖咏梅. 林缘和荒草坡不同草本层盖度小生境中积雪草的等级可塑性[J]. 植物生态学报, 2007, 31(4): 576-587. |
| [11] | 沈栋伟, 李媛媛, 陈小勇. 植物克隆多样性与生态系统功能[J]. 植物生态学报, 2007, 31(4): 552-560. |
| [12] | 蔡颖, 关保华, 安树青, 申瑞玲, 蒋金辉, 董蕾. 克隆植物乌菱对底泥磷含量及植株密度的表型可塑性响应[J]. 植物生态学报, 2007, 31(4): 599-606. |
| [13] | 魏晓慧, 殷东生, 祝宁. 自然条件下风箱果的克隆构型[J]. 植物生态学报, 2007, 31(4): 625-629. |
| [14] | 张淑敏, 于飞海, 董鸣. 土壤养分水平影响绢毛匍匐委陵菜匍匐茎生物量投资[J]. 植物生态学报, 2007, 31(4): 652-657. |
| [15] | 魏宇昆, 高玉葆, 李川, 许华, 任安芝. 内蒙古中东部草原羽茅内生真菌的遗传多样性[J]. 植物生态学报, 2006, 30(4): 640-649. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
Copyright © 2026 版权所有 《植物生态学报》编辑部
地址: 北京香山南辛村20号, 邮编: 100093
Tel.: 010-62836134, 62836138; Fax: 010-82599431; E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
备案号: 京ICP备16067583号-19
![]()