

Chin J Plant Ecol ›› 2021, Vol. 45 ›› Issue (9): 987-995.DOI: 10.17521/cjpe.2020.0366
• Research Articles • Previous Articles Next Articles
WANG Chun-Cheng1, ZHANG Yun-Ling2, MA Song-Mei3,*( ), HUANG Gang1, ZHANG Dan1, YAN Han3
), HUANG Gang1, ZHANG Dan1, YAN Han3
Received:2020-11-09
Accepted:2021-06-02
Online:2021-09-20
Published:2021-11-18
Contact:
MA Song-Mei
Supported by:WANG Chun-Cheng, ZHANG Yun-Ling, MA Song-Mei, HUANG Gang, ZHANG Dan, YAN Han. Phylogeny and species differentiation of four wild almond species of subgen. Amygdalus in China[J]. Chin J Plant Ecol, 2021, 45(9): 987-995.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.plant-ecology.com/EN/10.17521/cjpe.2020.0366
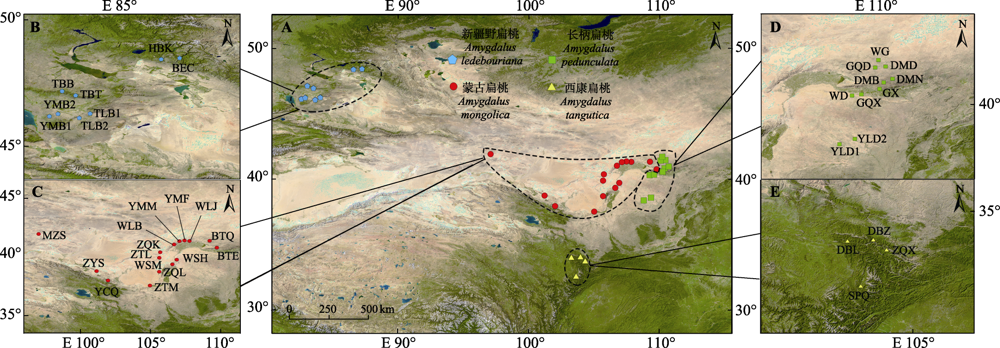
Fig. 1 Field sampling points of four subgen. Amygdalus wild plants. BEC, Buerjin County; BTE, Group 2 in Daqing Mountain; BTQ, Group 1 in Daqing Mountain; DBL, Group 1 in Têwo County; DBZ, Group 2 in Têwo County; DMB, Group 3 in Damao Banner; DMD, Group 1 in Damao Banner; DMN, Group 2 in Damao Banner; GQD, Group 1 in Guyang County; GQX, Group 3 in Guyang County; GX, Group 2 in Guyang County; HBK, Habahe County; MZS, Mazong Mountain; SPQ, Songpan County; TBB, Group 2 Tacheng City; TBT, Group 1 Tacheng City; TLB1, Group 1 in Toli County; TLB2, Group 2 in Toli County; WD, Wula Mountain; WG, Ugai Sumu Township; WLB, Dam Mouth; WLJ, Wujiahe Town; WSH, Suhaitu; WSM, Damaili Furrow; YCQ, Longshou Mountain; YLD1, Group 1 in Yulin City; YLD2, Group 2 in Yulin City; YMB1, Group 1 in Yumin County; YMB2, Group 2 in Yumin County; YMF, Group 2 in Yinshan Mountains; YMM, Group 1 in Yinshan Mountains; ZQK, Tukemu Gazha; ZQL, Helan Mountain; ZQX, Zhugqu County; ZTL, Temowula Gazha; ZTM, Su Litu Gazha; ZYS, Qilianshan Mountain.
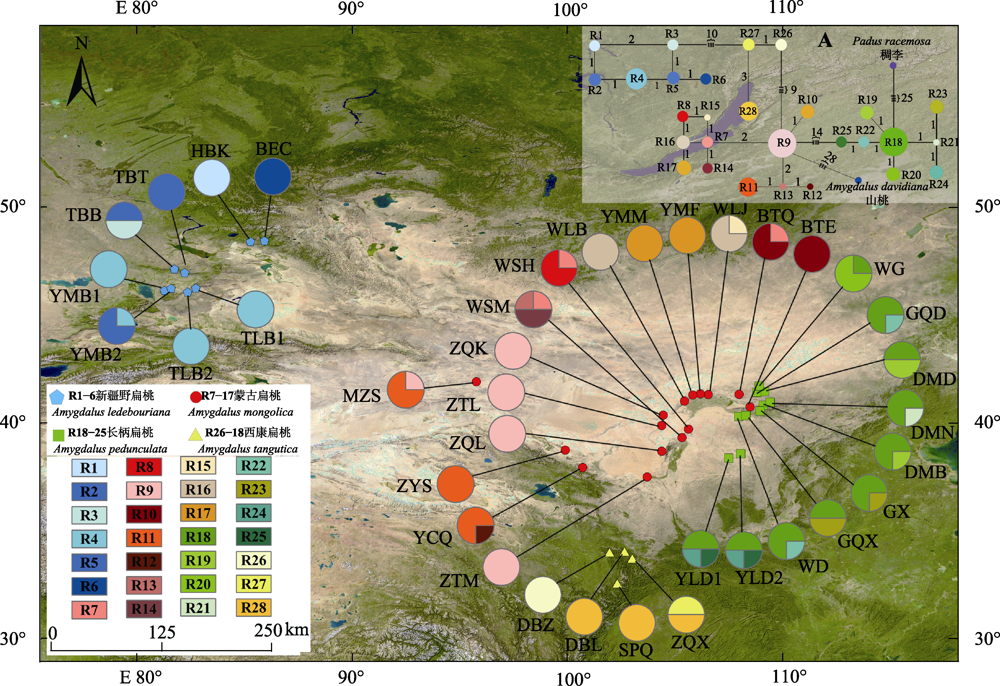
Fig. 2 Geographical distribution and the haplotype network of 28 haplotypes (R1-R28) of four subgen. Amygdalus wild plants. The population field sampling point codes in the figure are consistent with the ones in Fig. 1. Pie graphs indicate the frequency of each haplotype of these populations. A, In the median-joining haplotypes network, the sizes of the circles in the network are proportional to the haplotype frequencies. Branch lengths are roughly proportional to the number of mutation steps between haplotypes and nodes; the true number of steps is shown near the corresponding branch sections. Padus racemosa and Amygdalus davidiana was used as outgroup.
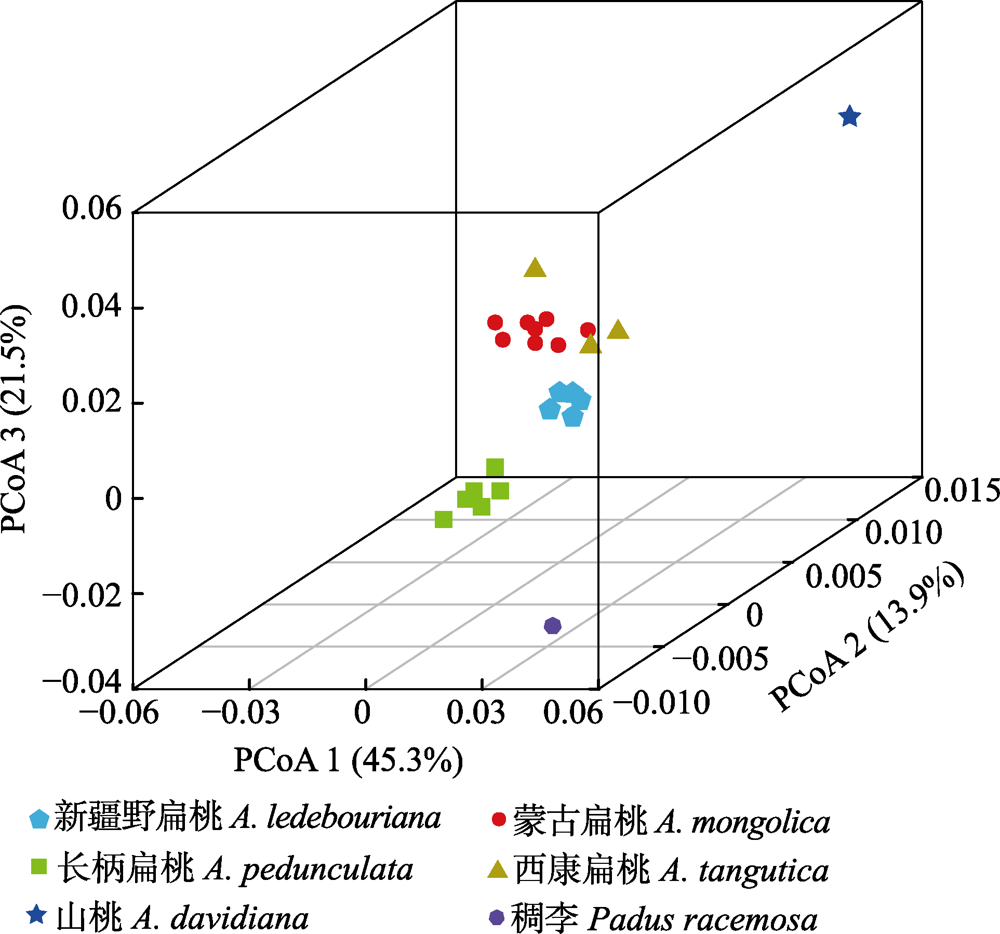
Fig. 4 Plots of the first three coordinates of the principal coordinates analysis (PCoA) at the population level for four subgen. Amygdalus wild plants.
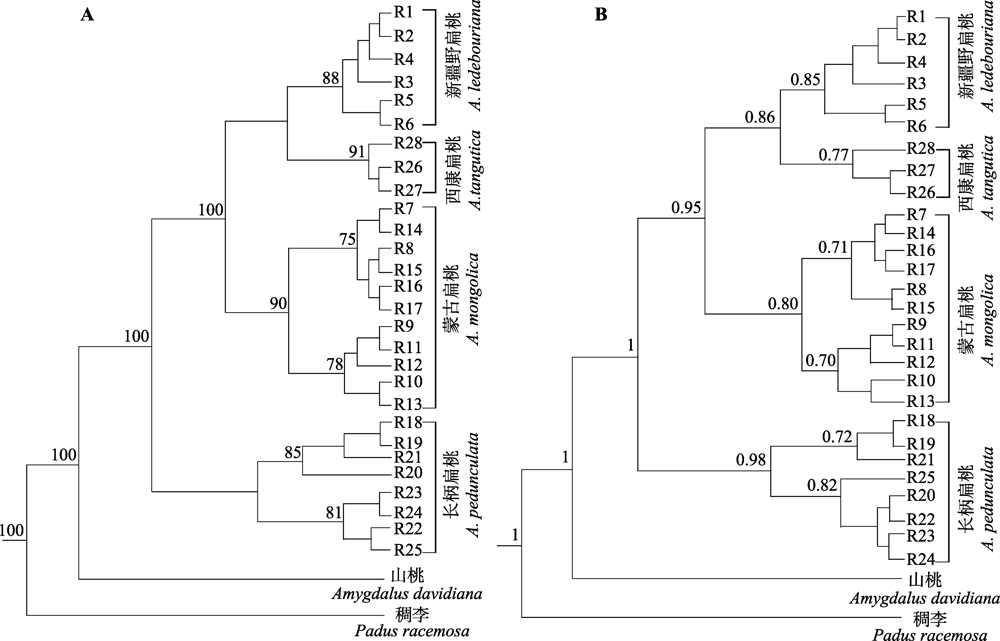
Fig. 5 Haplotype phylogenetic trees of four subgen. Amygdalus wild plants based on ITS sequences. A, Maximum likelihood (ML) tree. Bootstrap values equal to or greater than 70 are shown above the corresponding branching points. B, Bayesian tree. The values on the right of the branching points represent the posterior probability greater than 0.70; the brackets on the right of the two phylogenetic trees indicate the corresponding subclades of the four species.
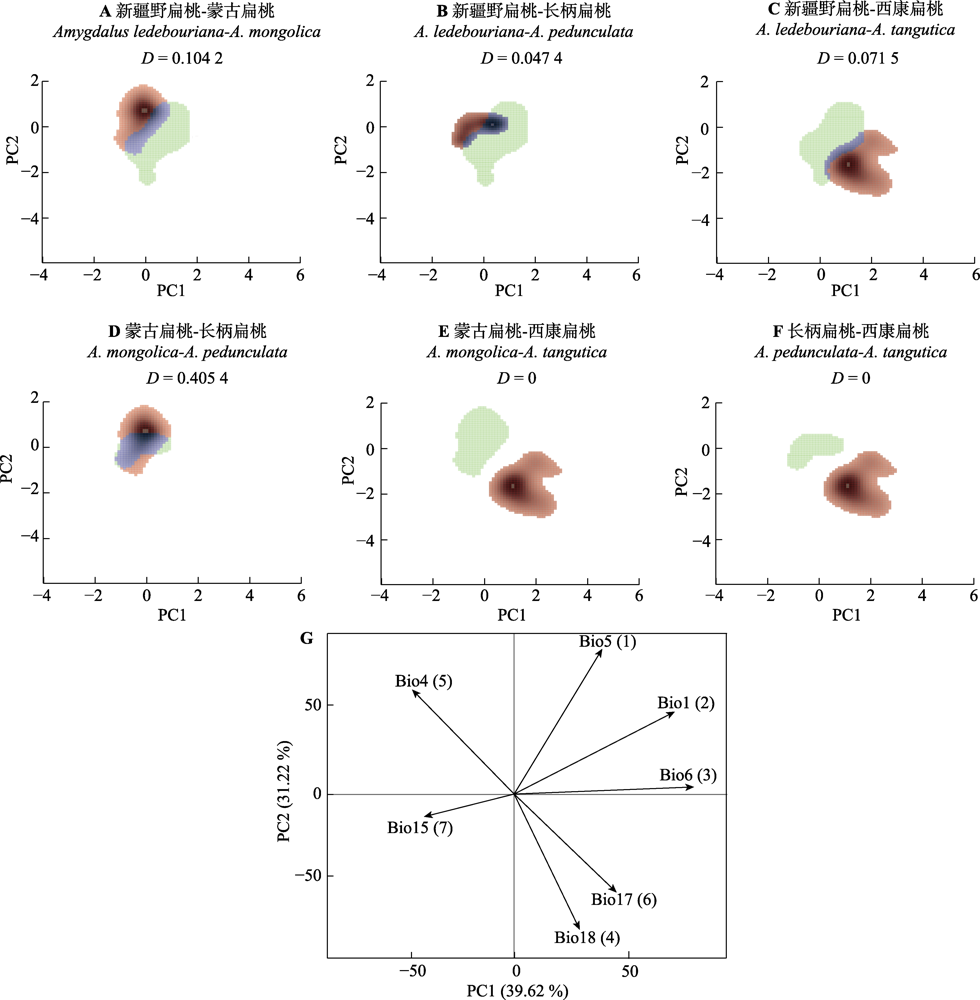
Fig. 6 Ecological niche differentiation and the contribution of driving factors of four subgen. Amygdalus plants. A-F, Visualization of ecological niches. The green colour depicts the niche space of the first species, red of the second species and the overlapping range is shown in blue. D is the ecological niche overlap score. G, Driver factors principal component analysis. The arrow depicts the direction of correlation (same direction indicates a high correlation). Bio1, annual mean temperature; Bio4, temperature seasonality (standard deviation × 100); Bio5, max temperature of warmest month; Bio6, min temperature of coldest month; Bio15, precipitation seasonality (coefficient of variation); Bio17, precipitation of driest quarter; Bio18, precipitation of warmest quarter. The numbers in brackets after the environmental factors are the contribution rankings.
| [1] | Bentham G, Hooker JD (1865). Genera Plantarum. A. Black, London. |
| [2] | Delplancke M, Yazbek M, Arrigo N, Espíndola A, Joly H, Alvarez N (2016). Combining conservative and variable markers to infer the evolutionary history of Prunus subgen. Amygdalus s.l. under domestication. Genetic Resources & Crop Evolution, 63, 221-234. |
| [3] | Dong SP (2015). Phylogeny Research of Prunus triloba and Related Species Based on Chromosome Kayotype and Single-copy Nuclear Gene DNA Sequences. Master degree dissertation, Beijing Forestry University, Beijing. |
| [ 董山平 (2015). 基于染色体核型和单拷贝核基因DNA序列的榆叶梅及其近缘种系统学研究. 硕士学位论文, 北京林业大学, 北京.] | |
| [4] | Duan YZ, Wang JH, Wang C, Wang HT, Du ZY (2020). Analysis on the potential suitable areas of four species of subgen. Amygdalus in arid Northwest China under future climate change. Chinese Journal of Ecology, 39, 2193-2204. |
| [ 段义忠, 王佳豪, 王驰, 王海涛, 杜忠毓 (2020). 未来气候变化下西北干旱区4种扁桃亚属植物潜在适生区分析. 生态学杂志, 39, 2193-2204.] | |
| [5] | Hall TA (1999). BioEdit:a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95-98. |
| [6] |
Jing ZB, Cheng JM, Guo CH, Wang XP (2013). Seed traits, nutrient elements and assessment of genetic diversity for almond (Amygdalus spp.) endangered to China as revealed using SRAP markers. Biochemical Systematics and Ecology, 49, 51-57.
DOI URL |
| [7] | Li YX (2001). Evolution of plant cell karyotype. Bulletin of Biology, 36(2), 16-17. |
| [ 李雅轩 (2001). 植物细胞核型的进化. 生物学通报, 36(2), 16-17.] | |
| [8] |
Librado P, Rozas J (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451-1452.
DOI PMID |
| [9] | Liu N, Huang Y (2010). Advances in molecular systematics of polyneoptera. Entomotaxonomia, 32, 304-312. |
| [ 刘念, 黄原 (2010). 多新翅类昆虫分子系统学的研究现状. 昆虫分类学报, 32, 304-312.] | |
| [10] |
Ma SM, Nie YB, Jiang XL, Xu Z, Ji WQ (2019). Genetic structure of the endangered, relict shrub Amygdalus mongolica (Rosaceae) in arid northwest China. Australian Journal of Botany, 67, 128-139.
DOI URL |
| [11] | Mao ZM, Li XY, Yang CY (1996). Flora of Xinjiang. Xinjiang Science, Technology and Health Press, Ürümqi. 355. |
| [ 毛祖美, 李学禹, 杨昌友 (1996). 新疆植物志. 新疆科技卫生出版社, 乌鲁木齐. 355.] | |
| [12] | Mei LX, Liu WQ, Wei Y, Jiang B (2014). Evaluation of the resources and development potential of Amygdalus spp. in China. Journal of Northwest Forestry University, 29(1), 69-72. |
| [ 梅立新, 刘文倩, 魏钰, 蒋宝 (2014). 中国扁桃资源与利用价值分析. 西北林学院学报, 29(1), 69-72.] | |
| [13] |
Peakall R, Smouse PE (2006). Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6, 288-295.
DOI URL |
| [14] |
Peterson AT (1999). Conservatism of ecological niches in evolutionary time. Science, 285, 1265-1267.
PMID |
| [15] |
Peterson AT, Soberón J, Sánchez CV (1999). Conservatism of ecological niches in evolutionary time. Science, 285, 1265-1267.
PMID |
| [16] | Qiu R, Cheng ZP, Wang ZL (2012). Studies on genetic relationship and evolutionary path of subgenus Amygdalus in China. Acta Horticulturae Sinica, 39, 205-214. |
| [ 邱蓉, 程中平, 王章利 (2012). 中国扁桃亚属植物亲缘关系及其演化途径研究. 园艺学报, 39, 205-214.] | |
| [17] |
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539-542.
DOI PMID |
| [18] | Shi WT (2018). Classification of Prunus L. sensu lato Based on Fruit Morphology. Master degree dissertation, South China Agricultural University, Guangdong. |
| [ 石文婷 (2018). 基于果实形态的广义李属植物系统分类学研究. 硕士学位论文, 华南农业大学, 广东.] | |
| [19] |
Simmons MP, Ochoterena H (2000). Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology, 49, 369-381.
PMID |
| [20] |
Slatyer RA, Hirst M, Sexton JP (2013). Niche breadth predicts geographical range size: a general ecological pattern. Ecology Letters, 16, 1104-1114.
DOI PMID |
| [21] |
Stephens M, Donnelly P (2003). A comparison of Bayesian methods for haplotype reconstruction from population genotype data. The American Journal of Human Genetics, 73, 1162-1169.
DOI URL |
| [22] |
Tamura K, Dudley J, Nei M, Kumar S (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596- 1599.
DOI URL |
| [23] | Tian JB, He Y, Cheng EM (2008). Chinese Almond. China Agriculture Press, Beijing. 54-56. |
| [ 田建保, 何勇, 程恩明 (2008). 中国扁桃. 中国农业出版社, 北京. 54-56.] | |
| [24] |
Warren DL, Glor RE, Turelli M (2008). Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution, 62, 2868-2883.
DOI PMID |
| [25] | Wei Y, Guo CH, Zhang GQ, Meng QC, Guo GG (2012). Studies on cold-resistance of several Chinese almond species. Journal of Northwest A&F University (Natural Science Edition), 40(6), 99-106. |
| [ 魏钰, 郭春会, 张国庆, 孟庆超, 郭改改 (2012). 我国几个扁桃种抗寒性的研究. 西北农林科技大学学报(自然科学版), 40(6), 99-106.] | |
| [26] | Yang B, Meng QY, Zhang K, Duan YZ (2020). Chloroplast genome characterization and identification of genetic relationship of relict endangered plant Amygdalus nana. Bulletin of Botanical Research, 40, 686-695. |
| [ 杨斌, 孟庆瑶, 张凯, 段义忠 (2020). 孑遗濒危植物矮扁桃叶绿体全基因组特征分析及亲缘关系鉴定. 植物研究, 40, 686- 695.] | |
| [27] |
Yazbek M, Oh SH (2013). Peaches and almonds: phylogeny of Prunus subg. Amygdalus (Rosaceae) based on DNA sequences and morphology. Plant Systematics and Evolution, 299, 1403-1418.
DOI URL |
| [28] | Yu DJ (1986). Flora Reipublicae Popularis Sinicae. Science Press, Beijing. 11-17. |
| [ 俞德浚 (1986). 中国植物志. 科学出版社, 北京. 11-17.] | |
| [29] | Yu HH, Yang ZL, Yang X, Tang ZF, Shu X, Liu RN (2010). Advances in studies on ITS sequence of medicinal plants germplasm resources. Chinese Traditional & Herbal Drugs, 41, 491-496. |
| [ 于华会, 杨志玲, 杨旭, 谭梓峰, 舒枭, 刘若楠 (2010). 药用植物种质资源ITS序列研究进展. 中草药, 41, 419-496.] | |
| [30] | Zeng B (2008). A Study on Propagate Biological Characteristics and Genetic Diversity of Germplasm Resources of Amygdalus ledebouriana Schlecht. PhD dissertation, Xinjiang Agricultural University, Ürümqi. |
| [ 曾斌 (2008). 新疆野扁桃繁殖生物学特性及种质资源遗传多样性研究. 博士学位论文, 新疆农业大学, 乌鲁木齐.] | |
| [31] |
Zhang HX, Wang Q, Jia SW (2020). Genomic phylogeography of Gymnocarpos przewalskii (Caryophyllaceae): insights into habitat fragmentation in arid northwestern China. Diversity, 12, 335.
DOI URL |
| [32] | Zhang YJ, Li DZ (2011). Advances in phylogenomics based on complete chloroplast genomes. Plant Diversity and Resources, 33, 365-375. |
| [ 张韵洁, 李德铢 (2011). 叶绿体系统发育基因组学的研究进展. 植物分类与资源学报, 33, 365-375.] | |
| [33] | Zhao YZ (1995). Study on floristic geographical distribution of Amygdalus mongolica. Acta Scientiarum Naturalium Universitatis Neimongol, 26, 713-715. |
| [ 赵一之 (1995). 蒙古扁桃的植物区系地理分布研究. 内蒙古大学学报(自然科学版), 26, 713-715.] |
| [1] | CHEN Yu-Ting, MA Song-Mei, ZHANG Dan, ZHANG Lin, WANG Chun-Cheng. Diversity pattern and formation mechanism of sympatric Haloxylon ammodendron and Haloxylon persicum in Xinjiang, China [J]. Chin J Plant Ecol, 2024, 48(1): 56-67. |
| [2] | YANG Xin, REN Ming-Xun. Species distribution pattern and formation mechanism of mangrove plants around the South China Sea [J]. Chin J Plant Ecol, 2023, 47(8): 1105-1115. |
| [3] | XIANG Wei, HUANG Dong-Liu, ZHU Shi-Dan. Absorptive root anatomical traits of 26 tropical and subtropical fern species [J]. Chin J Plant Ecol, 2022, 46(5): 593-601. |
| [4] | YANG Ke-Tong, CHANG Hai-Long, CHEN Guo-Peng, YU Xiao-Ya, XIAN Jun-Ren. Stomatal traits of main greening plant species in Lanzhou [J]. Chin J Plant Ecol, 2021, 45(2): 187-196. |
| [5] | TANG Li-Li, ZHANG Mei, ZHAO Xiang-Lin, KANG Mu-Yi, LIU Hong-Yan, GAO Xian-Ming, YANG Tong, ZHENG Pu-Fan, SHI Fu-Chen. Species distribution and community assembly rules of Juglans mandshurica in North China [J]. Chin J Plant Ecol, 2019, 43(9): 753-761. |
| [6] | WANG Xue, CHEN Guang-Shui, YAN Xiao-Jun, CHEN Ting-Ting, JIANG Qi, CHEN Yu-Hui, FAN Ai-Lian, JIA Lin-Qiao, XIONG De-Cheng, HUANG Jin-Xue. Variations in the first-order root diameter in 89 woody species in a subtropical evergreen broadleaved forest [J]. Chin J Plant Ecol, 2019, 43(11): 969-978. |
| [7] | YANG Lei, SUN Han, FAN Yan-Wen, HAN Wei, ZENG Ling-Bing, LIU Chao, WANG Xiang-Ping. Changes in leaf nitrogen and phosphorus stoichiometry of woody plants along an altitudinal gradient in Changbai Mountain, China [J]. Chin J Plan Ecolo, 2017, 41(12): 1228-1238. |
| [8] | LU Ning-Na,ZHAO Zhi-Gang. Flower symmetry and flower size variability: an examination of Berg’s hypotheses in an alpine meadow [J]. Chin J Plant Ecol, 2014, 38(5): 460-467. |
| [9] | Jannathan MAMUT, TAN Dun-Yan. Gynomonoecy in angiosperms: phylogeny, sex expression and evolutionary significance [J]. Chin J Plant Ecol, 2014, 38(1): 76-90. |
| [10] | WANG Lu,LEI Yun,ZHANG Ming-Li. A preliminary study of molecular phylogeny and biogeography distribution pattern of Zelkova inferred from trnL-trnF and nrITS sequences [J]. Chin J Plant Ecol, 2013, 37(5): 407-414. |
| [11] | CHEN Yan-Song, ZHOU Shou-Biao, OU Zu-Lan, XU Zhong-Dong, HONG Xin. Seed mass variation in common plant species in Wanfoshan Natural Reservation Region, Anhui, China [J]. Chin J Plant Ecol, 2012, 36(8): 739-746. |
| [12] | TAN Dun-Yan, ZHANG Yang, WANG Ai-Bo. A review of geocarpy and amphicarpy in angiosperms, with special reference to their ecological adaptive significance [J]. Chin J Plant Ecol, 2010, 34(1): 72-88. |
| [13] | HE Ya-Ping, LIU Jian-Quan. A Review on Recent Advances in the Studies of Plant Breeding System [J]. Chin J Plant Ecol, 2003, 27(2): 151-163. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Chinese Journal of Plant Ecology
Tel: 010-62836134, 62836138, E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn