

植物生态学报 ›› 2021, Vol. 45 ›› Issue (9): 987-995.DOI: 10.17521/cjpe.2020.0366
王春成1, 张云玲2, 马松梅3,*( ), 黄刚1, 张丹1, 闫涵3
), 黄刚1, 张丹1, 闫涵3
收稿日期:2020-11-09
接受日期:2021-06-02
出版日期:2021-09-20
发布日期:2021-11-18
通讯作者:
马松梅
作者简介:ORCID: *马松梅: 0000-0002-3107-2256(shzmsm@126.com)
基金资助:
WANG Chun-Cheng1, ZHANG Yun-Ling2, MA Song-Mei3,*( ), HUANG Gang1, ZHANG Dan1, YAN Han3
), HUANG Gang1, ZHANG Dan1, YAN Han3
Received:2020-11-09
Accepted:2021-06-02
Online:2021-09-20
Published:2021-11-18
Contact:
MA Song-Mei
Supported by:摘要:
基于核基因ITS序列研究中国4种野生扁桃: 新疆野扁桃(Amygdalus ledebouriana)、蒙古扁桃(A. mongolica)、长柄扁桃(A. pedunculata)和西康扁桃(A. tangutica)的系统发育关系和物种分化, 为4种植物的遗传与演化研究提供数据支撑。利用单倍型网络和主坐标分析揭示单倍型的聚类关系; 利用最大似然树和贝叶斯系统树分析单倍型的系统发育关系; 利用R语言“ecospat”包分析4种扁桃的生态位分化及其环境驱动因子。4种扁桃ITS1-ITS4片段总长为634 bp, 鉴别出27个核苷酸变异位点, 共定义了28个单倍型。4种扁桃种间最小遗传距离均大于种内最大遗传距离, 种间存在显著的遗传分化。4种扁桃的单倍型聚为两支: 新疆野扁桃、蒙古扁桃和西康扁桃聚为一支, 长柄扁桃为另一支; 单倍型网络和主坐标分析揭示的单倍型聚类关系与系统树一致。西康扁桃与蒙古扁桃、与长柄扁桃之间均表现出了显著的生态位分化, 最暖月最高气温、年平均气温、最冷月最低气温和最暖季降水量是驱动物种生态位分化的关键因子。
王春成, 张云玲, 马松梅, 黄刚, 张丹, 闫涵. 中国扁桃亚属四种野生扁桃的系统发育与物种分化. 植物生态学报, 2021, 45(9): 987-995. DOI: 10.17521/cjpe.2020.0366
WANG Chun-Cheng, ZHANG Yun-Ling, MA Song-Mei, HUANG Gang, ZHANG Dan, YAN Han. Phylogeny and species differentiation of four wild almond species of subgen. Amygdalus in China. Chinese Journal of Plant Ecology, 2021, 45(9): 987-995. DOI: 10.17521/cjpe.2020.0366
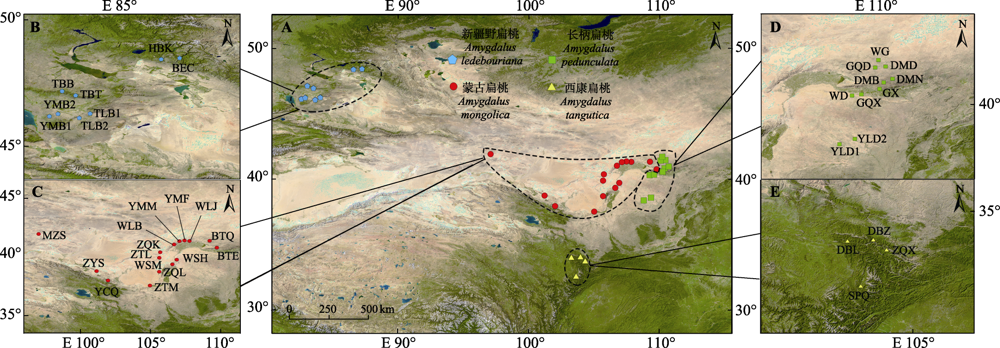
图1 4种扁桃亚属野生植物的野外采样点。BEC, 布尔津县; BTE, 大青山2; BTQ, 大青山1; DBL, 迭部县1; DBZ, 迭部县2; DMB, 达茂旗3; DMD, 达茂旗1; DMN, 达茂旗2; GQD, 固阳1; GQX, 固阳3; GX, 固阳2; HBK, 哈巴河县; MZS, 马鬃山; SPQ, 松潘县; TBB, 塔城2; TBT, 塔城1; TLB1, 托里县1; TLB2, 托里县2; WD, 乌拉山; WG, 乌盖苏木乡; WLB, 大坝口; WLJ, 乌加禾镇; WSH, 苏海图; WSM, 大麦力沟; YCQ, 龙首山; YLD1, 榆林1; YLD2, 榆林2; YMB1, 裕民县1; YMB2, 裕民县2; YMF, 阴山2; YMM, 阴山1; ZQK, 图克木嘎查; ZQL, 贺兰山; ZQX, 舟曲县; ZTL, 特莫乌拉嘎查; ZTM, 苏力图嘎查; ZYS, 祁连山。
Fig. 1 Field sampling points of four subgen. Amygdalus wild plants. BEC, Buerjin County; BTE, Group 2 in Daqing Mountain; BTQ, Group 1 in Daqing Mountain; DBL, Group 1 in Têwo County; DBZ, Group 2 in Têwo County; DMB, Group 3 in Damao Banner; DMD, Group 1 in Damao Banner; DMN, Group 2 in Damao Banner; GQD, Group 1 in Guyang County; GQX, Group 3 in Guyang County; GX, Group 2 in Guyang County; HBK, Habahe County; MZS, Mazong Mountain; SPQ, Songpan County; TBB, Group 2 Tacheng City; TBT, Group 1 Tacheng City; TLB1, Group 1 in Toli County; TLB2, Group 2 in Toli County; WD, Wula Mountain; WG, Ugai Sumu Township; WLB, Dam Mouth; WLJ, Wujiahe Town; WSH, Suhaitu; WSM, Damaili Furrow; YCQ, Longshou Mountain; YLD1, Group 1 in Yulin City; YLD2, Group 2 in Yulin City; YMB1, Group 1 in Yumin County; YMB2, Group 2 in Yumin County; YMF, Group 2 in Yinshan Mountains; YMM, Group 1 in Yinshan Mountains; ZQK, Tukemu Gazha; ZQL, Helan Mountain; ZQX, Zhugqu County; ZTL, Temowula Gazha; ZTM, Su Litu Gazha; ZYS, Qilianshan Mountain.
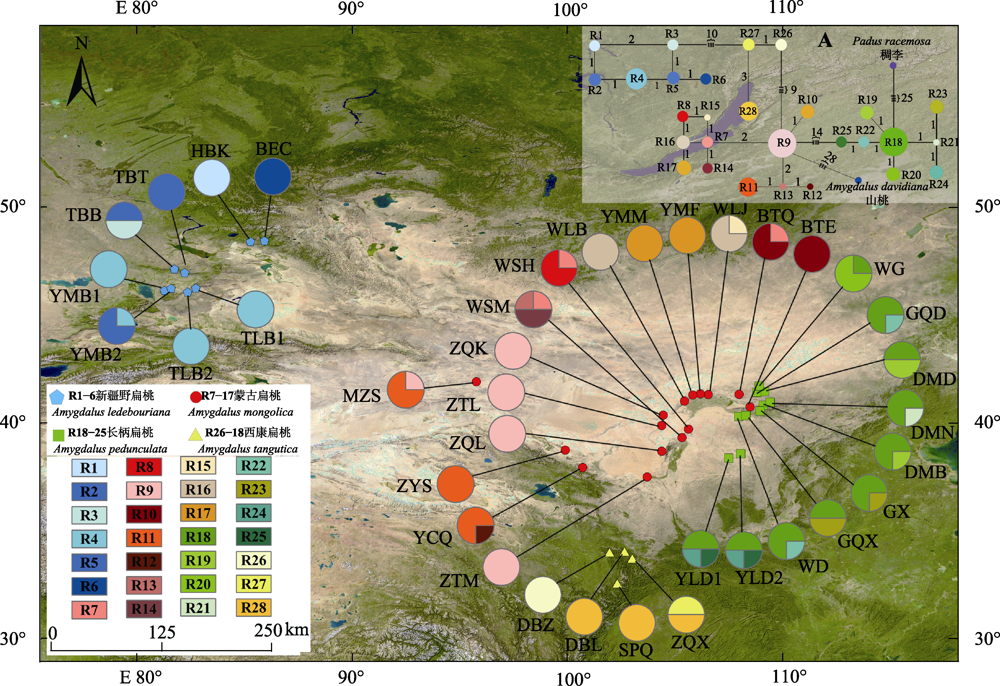
图2 扁桃亚属4种野生植物28个单倍型(R1-R28)的地理分布及其单倍型网络。图中种群采样点编码与图1一致, 饼图表示各种群的单倍型频率。A, 单倍型网络, 图中圆圈大小与单倍型频率成正比, 节点间的分支长度大致与单倍型的突变数成正比, 相应分支附近附有步长; 稠李和山桃作为外类群。
Fig. 2 Geographical distribution and the haplotype network of 28 haplotypes (R1-R28) of four subgen. Amygdalus wild plants. The population field sampling point codes in the figure are consistent with the ones in Fig. 1. Pie graphs indicate the frequency of each haplotype of these populations. A, In the median-joining haplotypes network, the sizes of the circles in the network are proportional to the haplotype frequencies. Branch lengths are roughly proportional to the number of mutation steps between haplotypes and nodes; the true number of steps is shown near the corresponding branch sections. Padus racemosa and Amygdalus davidiana was used as outgroup.
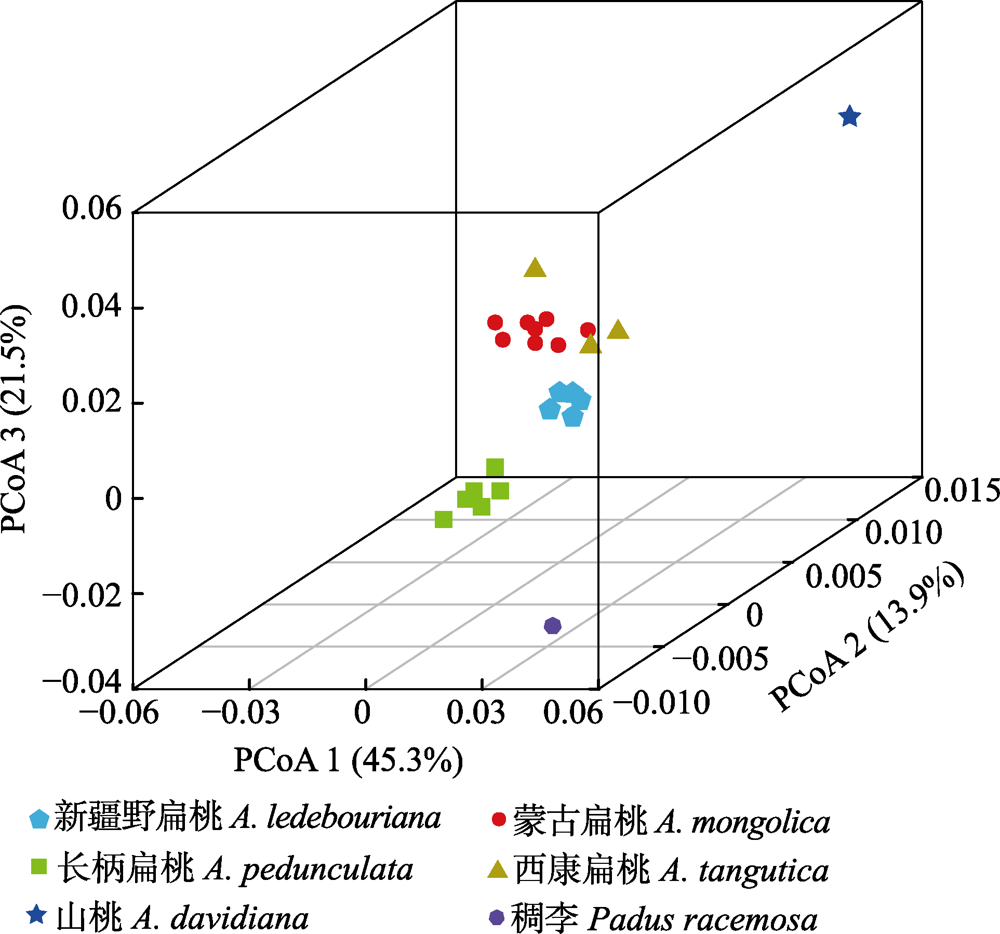
图4 扁桃亚属4种野生植物基于种群水平的前3个坐标的主坐标分析(PCoA)。
Fig. 4 Plots of the first three coordinates of the principal coordinates analysis (PCoA) at the population level for four subgen. Amygdalus wild plants.
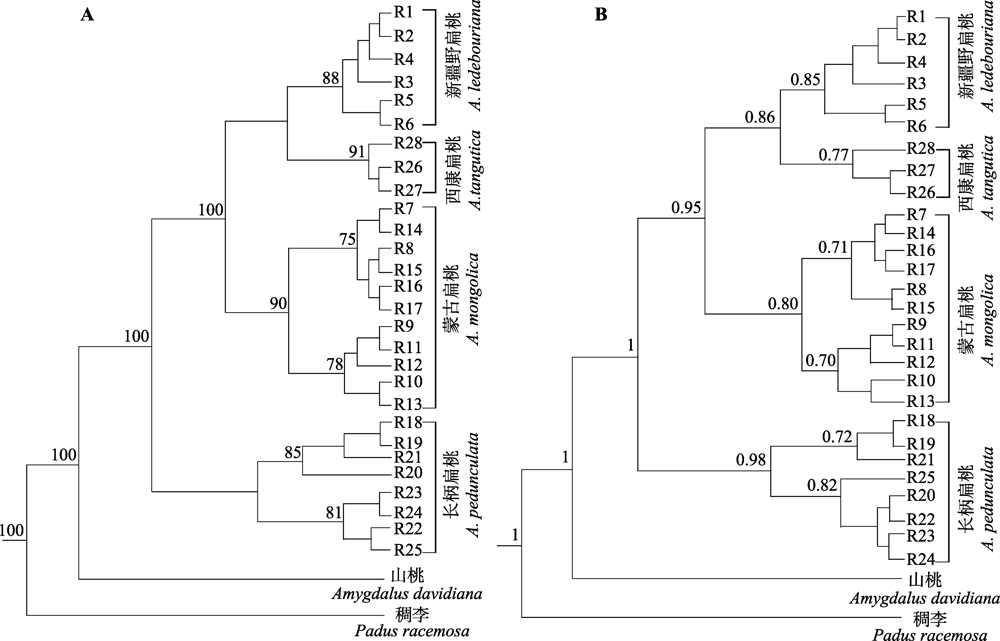
图5 扁桃亚属4种野生植物基于ITS序列的单倍型系统发育树。A, 最大似然(ML)树。分支点上方的数字为大于等于70的自展支持率。B, 贝叶斯树。分支节点右侧的数字表示大于0.70的后验概率值, 两系统发育树右侧的中括号表示为4个物种相应的亚支。
Fig. 5 Haplotype phylogenetic trees of four subgen. Amygdalus wild plants based on ITS sequences. A, Maximum likelihood (ML) tree. Bootstrap values equal to or greater than 70 are shown above the corresponding branching points. B, Bayesian tree. The values on the right of the branching points represent the posterior probability greater than 0.70; the brackets on the right of the two phylogenetic trees indicate the corresponding subclades of the four species.
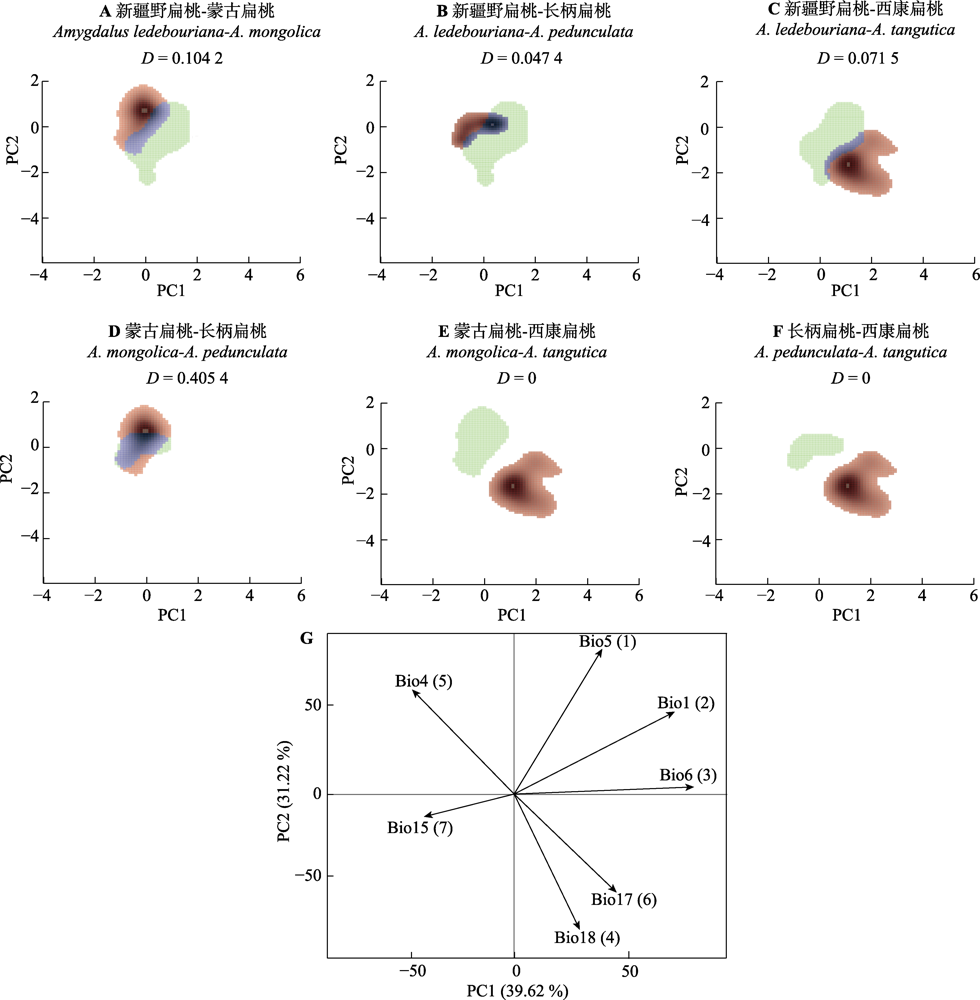
图6 扁桃亚属4种野生植物生态位相似性与驱动因子贡献率。A-F, 生态位可视化。绿色表示第一个种的生态位空间, 红色表示第二个种的生态位空间, 蓝色表示重叠空间。D为生态位重叠分数。G, 驱动因子主成分分析。箭头表示相关的方向(同一方向表示高度相关)。Bio1, 年平均气温; Bio4, 温度季节性变化标准差; Bio5, 最暖月最高气温; Bio6, 最冷月最低气温; Bio15, 降水量的季节性变异系数; Bio17, 最干季降水量; Bio18, 最暖季降水量。环境因子后括号里的数字为贡献率排名。
Fig. 6 Ecological niche differentiation and the contribution of driving factors of four subgen. Amygdalus plants. A-F, Visualization of ecological niches. The green colour depicts the niche space of the first species, red of the second species and the overlapping range is shown in blue. D is the ecological niche overlap score. G, Driver factors principal component analysis. The arrow depicts the direction of correlation (same direction indicates a high correlation). Bio1, annual mean temperature; Bio4, temperature seasonality (standard deviation × 100); Bio5, max temperature of warmest month; Bio6, min temperature of coldest month; Bio15, precipitation seasonality (coefficient of variation); Bio17, precipitation of driest quarter; Bio18, precipitation of warmest quarter. The numbers in brackets after the environmental factors are the contribution rankings.
| [1] | Bentham G, Hooker JD (1865). Genera Plantarum. A. Black, London. |
| [2] | Delplancke M, Yazbek M, Arrigo N, Espíndola A, Joly H, Alvarez N (2016). Combining conservative and variable markers to infer the evolutionary history of Prunus subgen. Amygdalus s.l. under domestication. Genetic Resources & Crop Evolution, 63, 221-234. |
| [3] | Dong SP (2015). Phylogeny Research of Prunus triloba and Related Species Based on Chromosome Kayotype and Single-copy Nuclear Gene DNA Sequences. Master degree dissertation, Beijing Forestry University, Beijing. |
| [ 董山平 (2015). 基于染色体核型和单拷贝核基因DNA序列的榆叶梅及其近缘种系统学研究. 硕士学位论文, 北京林业大学, 北京.] | |
| [4] | Duan YZ, Wang JH, Wang C, Wang HT, Du ZY (2020). Analysis on the potential suitable areas of four species of subgen. Amygdalus in arid Northwest China under future climate change. Chinese Journal of Ecology, 39, 2193-2204. |
| [ 段义忠, 王佳豪, 王驰, 王海涛, 杜忠毓 (2020). 未来气候变化下西北干旱区4种扁桃亚属植物潜在适生区分析. 生态学杂志, 39, 2193-2204.] | |
| [5] | Hall TA (1999). BioEdit:a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95-98. |
| [6] |
Jing ZB, Cheng JM, Guo CH, Wang XP (2013). Seed traits, nutrient elements and assessment of genetic diversity for almond (Amygdalus spp.) endangered to China as revealed using SRAP markers. Biochemical Systematics and Ecology, 49, 51-57.
DOI URL |
| [7] | Li YX (2001). Evolution of plant cell karyotype. Bulletin of Biology, 36(2), 16-17. |
| [ 李雅轩 (2001). 植物细胞核型的进化. 生物学通报, 36(2), 16-17.] | |
| [8] |
Librado P, Rozas J (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451-1452.
DOI PMID |
| [9] | Liu N, Huang Y (2010). Advances in molecular systematics of polyneoptera. Entomotaxonomia, 32, 304-312. |
| [ 刘念, 黄原 (2010). 多新翅类昆虫分子系统学的研究现状. 昆虫分类学报, 32, 304-312.] | |
| [10] |
Ma SM, Nie YB, Jiang XL, Xu Z, Ji WQ (2019). Genetic structure of the endangered, relict shrub Amygdalus mongolica (Rosaceae) in arid northwest China. Australian Journal of Botany, 67, 128-139.
DOI URL |
| [11] | Mao ZM, Li XY, Yang CY (1996). Flora of Xinjiang. Xinjiang Science, Technology and Health Press, Ürümqi. 355. |
| [ 毛祖美, 李学禹, 杨昌友 (1996). 新疆植物志. 新疆科技卫生出版社, 乌鲁木齐. 355.] | |
| [12] | Mei LX, Liu WQ, Wei Y, Jiang B (2014). Evaluation of the resources and development potential of Amygdalus spp. in China. Journal of Northwest Forestry University, 29(1), 69-72. |
| [ 梅立新, 刘文倩, 魏钰, 蒋宝 (2014). 中国扁桃资源与利用价值分析. 西北林学院学报, 29(1), 69-72.] | |
| [13] |
Peakall R, Smouse PE (2006). Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6, 288-295.
DOI URL |
| [14] |
Peterson AT (1999). Conservatism of ecological niches in evolutionary time. Science, 285, 1265-1267.
PMID |
| [15] |
Peterson AT, Soberón J, Sánchez CV (1999). Conservatism of ecological niches in evolutionary time. Science, 285, 1265-1267.
PMID |
| [16] | Qiu R, Cheng ZP, Wang ZL (2012). Studies on genetic relationship and evolutionary path of subgenus Amygdalus in China. Acta Horticulturae Sinica, 39, 205-214. |
| [ 邱蓉, 程中平, 王章利 (2012). 中国扁桃亚属植物亲缘关系及其演化途径研究. 园艺学报, 39, 205-214.] | |
| [17] |
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539-542.
DOI PMID |
| [18] | Shi WT (2018). Classification of Prunus L. sensu lato Based on Fruit Morphology. Master degree dissertation, South China Agricultural University, Guangdong. |
| [ 石文婷 (2018). 基于果实形态的广义李属植物系统分类学研究. 硕士学位论文, 华南农业大学, 广东.] | |
| [19] |
Simmons MP, Ochoterena H (2000). Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology, 49, 369-381.
PMID |
| [20] |
Slatyer RA, Hirst M, Sexton JP (2013). Niche breadth predicts geographical range size: a general ecological pattern. Ecology Letters, 16, 1104-1114.
DOI PMID |
| [21] |
Stephens M, Donnelly P (2003). A comparison of Bayesian methods for haplotype reconstruction from population genotype data. The American Journal of Human Genetics, 73, 1162-1169.
DOI URL |
| [22] |
Tamura K, Dudley J, Nei M, Kumar S (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596- 1599.
DOI URL |
| [23] | Tian JB, He Y, Cheng EM (2008). Chinese Almond. China Agriculture Press, Beijing. 54-56. |
| [ 田建保, 何勇, 程恩明 (2008). 中国扁桃. 中国农业出版社, 北京. 54-56.] | |
| [24] |
Warren DL, Glor RE, Turelli M (2008). Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution, 62, 2868-2883.
DOI PMID |
| [25] | Wei Y, Guo CH, Zhang GQ, Meng QC, Guo GG (2012). Studies on cold-resistance of several Chinese almond species. Journal of Northwest A&F University (Natural Science Edition), 40(6), 99-106. |
| [ 魏钰, 郭春会, 张国庆, 孟庆超, 郭改改 (2012). 我国几个扁桃种抗寒性的研究. 西北农林科技大学学报(自然科学版), 40(6), 99-106.] | |
| [26] | Yang B, Meng QY, Zhang K, Duan YZ (2020). Chloroplast genome characterization and identification of genetic relationship of relict endangered plant Amygdalus nana. Bulletin of Botanical Research, 40, 686-695. |
| [ 杨斌, 孟庆瑶, 张凯, 段义忠 (2020). 孑遗濒危植物矮扁桃叶绿体全基因组特征分析及亲缘关系鉴定. 植物研究, 40, 686- 695.] | |
| [27] |
Yazbek M, Oh SH (2013). Peaches and almonds: phylogeny of Prunus subg. Amygdalus (Rosaceae) based on DNA sequences and morphology. Plant Systematics and Evolution, 299, 1403-1418.
DOI URL |
| [28] | Yu DJ (1986). Flora Reipublicae Popularis Sinicae. Science Press, Beijing. 11-17. |
| [ 俞德浚 (1986). 中国植物志. 科学出版社, 北京. 11-17.] | |
| [29] | Yu HH, Yang ZL, Yang X, Tang ZF, Shu X, Liu RN (2010). Advances in studies on ITS sequence of medicinal plants germplasm resources. Chinese Traditional & Herbal Drugs, 41, 491-496. |
| [ 于华会, 杨志玲, 杨旭, 谭梓峰, 舒枭, 刘若楠 (2010). 药用植物种质资源ITS序列研究进展. 中草药, 41, 419-496.] | |
| [30] | Zeng B (2008). A Study on Propagate Biological Characteristics and Genetic Diversity of Germplasm Resources of Amygdalus ledebouriana Schlecht. PhD dissertation, Xinjiang Agricultural University, Ürümqi. |
| [ 曾斌 (2008). 新疆野扁桃繁殖生物学特性及种质资源遗传多样性研究. 博士学位论文, 新疆农业大学, 乌鲁木齐.] | |
| [31] |
Zhang HX, Wang Q, Jia SW (2020). Genomic phylogeography of Gymnocarpos przewalskii (Caryophyllaceae): insights into habitat fragmentation in arid northwestern China. Diversity, 12, 335.
DOI URL |
| [32] | Zhang YJ, Li DZ (2011). Advances in phylogenomics based on complete chloroplast genomes. Plant Diversity and Resources, 33, 365-375. |
| [ 张韵洁, 李德铢 (2011). 叶绿体系统发育基因组学的研究进展. 植物分类与资源学报, 33, 365-375.] | |
| [33] | Zhao YZ (1995). Study on floristic geographical distribution of Amygdalus mongolica. Acta Scientiarum Naturalium Universitatis Neimongol, 26, 713-715. |
| [ 赵一之 (1995). 蒙古扁桃的植物区系地理分布研究. 内蒙古大学学报(自然科学版), 26, 713-715.] |
| [1] | 陈雨婷, 马松梅, 张丹, 张林, 王春成. 新疆同域分布梭梭和白梭梭多样性格局及其形成机制[J]. 植物生态学报, 2024, 48(1): 56-67. |
| [2] | 杨鑫, 任明迅. 环南海区域红树物种多样性分布格局及其形成机制[J]. 植物生态学报, 2023, 47(8): 1105-1115. |
| [3] | 项伟, 黄冬柳, 朱师丹. 热带亚热带26种蕨类植物的吸收根解剖特征[J]. 植物生态学报, 2022, 46(5): 593-601. |
| [4] | 杨克彤, 常海龙, 陈国鹏, 俞筱押, 鲜骏仁. 兰州市主要绿化植物气孔性状特征[J]. 植物生态学报, 2021, 45(2): 187-196. |
| [5] | 闫涵, 张云玲, 马松梅, 王春成, 张丹. 黑果枸杞在新疆的适宜分布模拟与局部环境适应性分化[J]. 植物生态学报, 2021, 45(11): 1221-1230. |
| [6] | 柴永福, 许金石, 刘鸿雁, 刘全儒, 郑成洋, 康慕谊, 梁存柱, 王仁卿, 高贤明, 张峰, 福臣, 刘晓, 岳明. 华北地区主要灌丛群落物种组成及系统发育结构特征[J]. 植物生态学报, 2019, 43(9): 793-805. |
| [7] | 闫雅楠, 叶小齐, 吴明, 闫明, 张昕丽. 入侵植物加拿大一枝黄花根际解钾菌多样性及解钾活性[J]. 植物生态学报, 2019, 43(6): 543-556. |
| [8] | 王雪, 陈光水, 闫晓俊, 陈廷廷, 姜琦, 陈宇辉, 范爱连, 贾林巧, 熊德成, 黄锦学. 亚热带常绿阔叶林89种木本植物一级根直径的变异[J]. 植物生态学报, 2019, 43(11): 969-978. |
| [9] | 赵乐文, 陈梓熠, 邹滢, 付子钊, 吴桂林, 刘小容, 罗琦, 林忆雪, 李雄炬, 刘智通, 刘慧. 九种维管植物水力性状的演化趋势[J]. 植物生态学报, 2018, 42(2): 220-228. |
| [10] | 车应弟, 刘旻霞, 李俐蓉, 焦骄, 肖卫. 基于功能性状及系统发育的亚高寒草甸群落构建[J]. 植物生态学报, 2017, 41(11): 1157-1167. |
| [11] | 姜超, 谭珂, 任明迅. 季风对亚洲热带植物分布格局的影响[J]. 植物生态学报, 2017, 41(10): 1103-1112. |
| [12] | 路宁娜,赵志刚. 花对称性与植物花大小的变异性: 在高寒草甸植物群落检验Berg的假说[J]. 植物生态学报, 2014, 38(5): 460-467. |
| [13] | 邓建明, 姚步青, 周华坤, 赵新全, 魏晴, 陈哲, 王文颖. 水氮添加条件下高寒草甸主要植物种氮素吸收分配的同位素示踪研究[J]. 植物生态学报, 2014, 38(2): 116-124. |
| [14] | 王璐,雷耘,张明理. 基于序列trnL-trnF和ITS的榉属系统发育与地理分布格局的初步分析[J]. 植物生态学报, 2013, 37(5): 407-414. |
| [15] | 陈延松, 周守标, 欧祖兰, 徐忠东, 洪欣. 安徽万佛山自然保护区常见植物种子大小变异[J]. 植物生态学报, 2012, 36(8): 739-746. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
Copyright © 2022 版权所有 《植物生态学报》编辑部
地址: 北京香山南辛村20号, 邮编: 100093
Tel.: 010-62836134, 62836138; Fax: 010-82599431; E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
备案号: 京ICP备16067583号-19