

植物生态学报 ›› 2024, Vol. 48 ›› Issue (2): 215-228.DOI: 10.17521/cjpe.2022.0433 cstr: 32100.14.cjpe.2022.0433
程可心, 杜尧, 李凯航, 王浩臣, 杨艳, 金一*( ), 何晓青*(
), 何晓青*( )
)
收稿日期:2022-10-31
接受日期:2023-05-16
出版日期:2024-02-28
发布日期:2023-05-17
通讯作者:
* (基金资助:
CHENG Ke-Xin, DU Yao, LI Kai-Hang, WANG Hao-Chen, YANG Yan, JIN Yi*( ), HE Xiao-Qing*(
), HE Xiao-Qing*( )
)
Received:2022-10-31
Accepted:2023-05-16
Online:2024-02-28
Published:2023-05-17
Contact:
* (Supported by:摘要:
为了解玉米(Zea mays)和其定植微生物组之间的相互作用, 探究玉米与叶际微生物组之间的互作遗传机制, 该研究采用数学模型量化微生物之间相互作用的4种方式: 互利共生、拮抗、侵略、利他, 分析230份玉米叶际微生物组数据, 利用网络作图研究玉米与叶际微生物组之间的互作遗传机制。结果表明: 在微生物互作网络中确定了67个中心节点微生物, 通过网络作图筛选到玉米405个显著单核苷酸多态性(SNPs)位点, 最终定位到23个枢纽基因, 发现其在促进植物生长、抵御病原菌侵染、耐受非生物胁迫方面起到重要作用。研究结果有助于在作物遗传育种以及构建新型菌剂促进植物生长方面提供思路。
程可心, 杜尧, 李凯航, 王浩臣, 杨艳, 金一, 何晓青. 玉米与叶际微生物组的互作遗传机制. 植物生态学报, 2024, 48(2): 215-228. DOI: 10.17521/cjpe.2022.0433
CHENG Ke-Xin, DU Yao, LI Kai-Hang, WANG Hao-Chen, YANG Yan, JIN Yi, HE Xiao-Qing. Genetic mechanism of interaction between maize and phyllospheric microbiome. Chinese Journal of Plant Ecology, 2024, 48(2): 215-228. DOI: 10.17521/cjpe.2022.0433
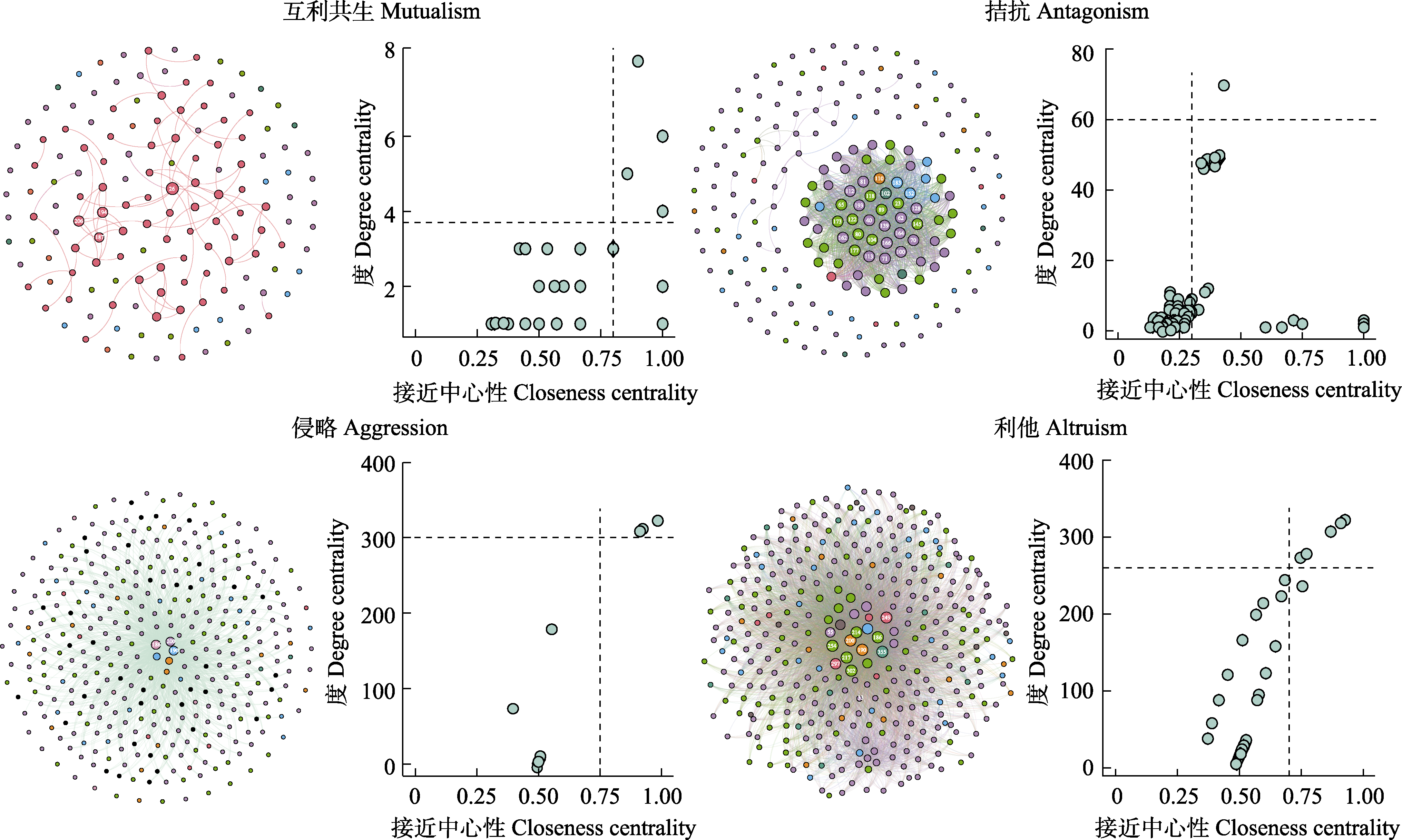
图1 玉米叶际微生物在4种互作关系网络下的中心节点微生物。在4个不同的共现网络中, 中心节点微生物是基于特征值度和接近中心性进行划分的, 每个网络的右侧显示分类操作单元(OTU)的这两个值, 进行中心节点微生物的筛选, 并把中心节点微生物在网络中用不同的数字进行表示。OTU对应的物种组成见表1, 最终使用Gephi软件对4种互作关系进行可视化。
Fig. 1 Hub microbes of maize phyllosphere microorganisms under the four different co-occurrence networks. In four different co-occurrence networks, hub microbes are divided based on degree centrality and closeness centrality, the right side of each network shows these two values of the operational taxonomic unit (OTU). The hub microbes are represented by different numbers in the network, the species composition corresponding to OTU is shown in Table 1, and finally the four interaction relationships are visualized using Gephi software.
| OTU名称 OTU name | 门 Dividion | 纲 Class | 目 Order | 科 Family | 属 Genus | |
|---|---|---|---|---|---|---|
| 互利共生 | 4 | 变形菌门 | α变形菌纲 | 根瘤菌目 Rhizobiales | 甲基杆菌科 | 甲基杆菌属 |
| Mutualism | Proteobacteria | Alphaproteobacteria | Methylobacteriaceae | Methylobacterium | ||
| 26 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 194 | 放线菌门 | 放线菌纲 | NA | NA | NA | |
| Actinobacteria | Actinobacteria | |||||
| 206 | 放线菌门 | 放线菌纲 | 放线菌目 | NA | NA | |
| Actinobacteria | Actinobacteria | Actinomycetales | ||||
| 187 | 放线菌门 | NA | NA | NA | NA | |
| Actinobacteria | ||||||
| 拮抗 | 23 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 |
| Antagonism | Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | |
| 40 | 放线菌门 | 放线菌纲 | 放线菌目 | 动孢囊菌科 | NA | |
| Actinobacteria | Actinobacteria | Actinomycetales | Kineosporiaceae | |||
| 62 | 拟杆菌门 | Sphingobacteriia | Sphingobacteriales | Sphingobacteriaceae | 土地杆菌属 | |
| Bacteroidetes | Pedobacter | |||||
| 65 | 放线菌门 | 放线菌纲 | 放线菌目 | 动孢囊菌科 | 动球菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Kineosporiaceae | Kineococcus | ||
| 70 | 放线菌门 | 放线菌纲 | 放线菌目 | 微杆菌科 | 寒冷杆菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Frigoribacterium | ||
| 71 | 放线菌门 | 放线菌纲 | 放线菌目 | 微杆菌科 | 拉氏拉氏杆菌 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Rathayibacter | ||
| 80 | 变形菌门 | α变形菌纲 | 根瘤菌目 | 甲基杆菌科 | 甲基杆菌属 | |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | Methylobacterium | ||
| 81 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 89 | 放线菌门 | 放线菌纲 | 放线菌目 | 微杆菌科 | 微杆菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Microbacterium | ||
| 100 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 102 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 104 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 110 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 112 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 113 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 118 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 122 | 放线菌门 | 放线菌纲 | 放线菌目 | 微球菌科 | 微杆菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Microbacterium | ||
| 128 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 139 | 变形菌门 | α变形菌纲 | 根瘤菌目 | 甲基杆菌科 | 甲基杆菌属 | |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | Methylobacterium | ||
| 145 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 152 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 162 | 放线菌门 | 放线菌纲 | 放线菌目 | 微球菌科 | 微杆菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Microbacterium | ||
| 164 | 放线菌门 | 放线菌纲 | 放线菌目 | 微球菌科 | 微杆菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Microbacterium | ||
| 166 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 173 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 177 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 183 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 193 | 放线菌门 | 放线菌纲 | NA | NA | NA | |
| Actinobacteria | Acidimicrobiia | |||||
| 205 | 放线菌门 | 放线菌纲 | 放线菌目 | NA | NA | |
| Actinobacteria | Acidimicrobiia | Acidimicrobiales | ||||
| 207 | 放线菌门 | 嗜热油菌纲 | 索利红杆菌目 | NA | NA | |
| Actinobacteria | Thermoleophilia | Solirubrobacterales | ||||
| 212 | 厚壁菌门 | 芽孢杆菌纲 | 乳杆菌目 | NA | NA | |
| Firmicutes | Bacilli | Lactobacillales | ||||
| 218 | 变形菌门 | β变形菌纲 | NA | NA | NA | |
| Proteobacteria | Betaproteobacteria | |||||
| 233 | 放线菌门 | 放线菌纲 | 放线菌目 | 诺卡氏菌科 | NA | |
| Actinobacteria | Actinobacteria | Actinomycetales | Nocardiaceae | |||
| 253 | 变形菌门 | α变形菌纲 | 红螺菌目 | 红螺菌科 | NA | |
| Proteobacteria | Alphaproteobacteria | Rhodospirillales | Rhodospirillaceae | |||
| 255 | 变形菌门 | β变形菌纲 | NA | NA | NA | |
| Proteobacteria | Betaproteobacteria | |||||
| 271 | 放线菌门 | 放线菌纲 | 放线菌目 | 动孢囊菌科 | 动球菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Kineosporiaceae | Kineococcus | ||
| 274 | 放线菌门 | 放线菌纲 | 放线菌目 | 微球菌科 | 寒冷杆菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Frigoribacterium | ||
| 278 | 放线菌门 | 放线菌纲 | 放线菌目 | 诺卡氏菌科 | 马红球菌 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Nocardiaceae | Rhodococcus | ||
| 301 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | Kaistobacter | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | |||
| 303 | 变形菌门 | β变形菌纲 | NA | NA | NA | |
| Proteobacteria | Betaproteobacteria | |||||
| 313 | 变形菌门 | γ变形菌纲 | 假单胞菌目 | 假单胞菌科 | NA | |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | |||
| 323 | 放线菌门 | 放线菌纲 | 放线菌目 | 动孢囊菌科 | 动球菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Kineosporiaceae | Kineococcus | ||
| 326 | 放线菌门 | 放线菌纲 | 放线菌目 | 微杆菌科 | 寒冷杆菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Frigoribacterium | ||
| 354 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | Kaistobacter | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | |||
| 359 | 变形菌门 | β变形菌纲 | NA | NA | NA | |
| Proteobacteria | Betaproteobacteria | |||||
| 369 | 变形菌门 | γ变形菌纲 | 假单胞菌目 | 莫拉氏菌属 | 不动杆菌属 | |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | Acinetobacter | ||
| 370 | 变形菌门 | γ变形菌纲 | 假单胞菌目 | 假单胞菌科 | NA | |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | |||
| 侵略 | 186 | NA | NA | NA | NA | NA |
| Aggression | 190 | 变形菌门 | NA | NA | NA | NA |
| Proteobacteria | ||||||
| 200 | 变形菌门 | α变形菌纲 | NA | NA | NA | |
| Proteobacteria | Alphaproteobacteria | |||||
| 利他 | 186 | NA | NA | NA | NA | NA |
| Altruism | 190 | 变形菌门 | NA | NA | NA | NA |
| Proteobacteria | ||||||
| 200 | 变形菌门 | α变形菌纲 | NA | NA | NA | |
| Proteobacteria | Alphaproteobacteria | |||||
| 217 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | NA | NA | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | ||||
| 254 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | NA | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | |||
| 302 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 355 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 214 | 变形菌门 | α变形菌纲 | 根瘤菌目 | NA | NA | |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | ||||
| 249 | 变形菌门 | α变形菌纲 | 根瘤菌目 | 甲基杆菌科 | NA | |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | |||
| 297 | 变形菌门 | α变形菌纲 | 根瘤菌目 | 甲基杆菌科 | 甲基杆菌属 | |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | Methylobacterium | ||
| 55 | 变形菌门 | α变形菌纲 | 根瘤菌目 | 甲基杆菌科 | 甲基杆菌属 | |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | Methylobacterium | ||
| 33 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas |
表1 四种互作网络的中心节点微生物可操作分类单元(OTU)
Table 1 List of ecologically hub Operational Taxonomic Units (OTU)
| OTU名称 OTU name | 门 Dividion | 纲 Class | 目 Order | 科 Family | 属 Genus | |
|---|---|---|---|---|---|---|
| 互利共生 | 4 | 变形菌门 | α变形菌纲 | 根瘤菌目 Rhizobiales | 甲基杆菌科 | 甲基杆菌属 |
| Mutualism | Proteobacteria | Alphaproteobacteria | Methylobacteriaceae | Methylobacterium | ||
| 26 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 194 | 放线菌门 | 放线菌纲 | NA | NA | NA | |
| Actinobacteria | Actinobacteria | |||||
| 206 | 放线菌门 | 放线菌纲 | 放线菌目 | NA | NA | |
| Actinobacteria | Actinobacteria | Actinomycetales | ||||
| 187 | 放线菌门 | NA | NA | NA | NA | |
| Actinobacteria | ||||||
| 拮抗 | 23 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 |
| Antagonism | Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | |
| 40 | 放线菌门 | 放线菌纲 | 放线菌目 | 动孢囊菌科 | NA | |
| Actinobacteria | Actinobacteria | Actinomycetales | Kineosporiaceae | |||
| 62 | 拟杆菌门 | Sphingobacteriia | Sphingobacteriales | Sphingobacteriaceae | 土地杆菌属 | |
| Bacteroidetes | Pedobacter | |||||
| 65 | 放线菌门 | 放线菌纲 | 放线菌目 | 动孢囊菌科 | 动球菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Kineosporiaceae | Kineococcus | ||
| 70 | 放线菌门 | 放线菌纲 | 放线菌目 | 微杆菌科 | 寒冷杆菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Frigoribacterium | ||
| 71 | 放线菌门 | 放线菌纲 | 放线菌目 | 微杆菌科 | 拉氏拉氏杆菌 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Rathayibacter | ||
| 80 | 变形菌门 | α变形菌纲 | 根瘤菌目 | 甲基杆菌科 | 甲基杆菌属 | |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | Methylobacterium | ||
| 81 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 89 | 放线菌门 | 放线菌纲 | 放线菌目 | 微杆菌科 | 微杆菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Microbacterium | ||
| 100 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 102 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 104 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 110 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 112 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 113 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 118 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 122 | 放线菌门 | 放线菌纲 | 放线菌目 | 微球菌科 | 微杆菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Microbacterium | ||
| 128 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 139 | 变形菌门 | α变形菌纲 | 根瘤菌目 | 甲基杆菌科 | 甲基杆菌属 | |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | Methylobacterium | ||
| 145 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 152 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 162 | 放线菌门 | 放线菌纲 | 放线菌目 | 微球菌科 | 微杆菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Microbacterium | ||
| 164 | 放线菌门 | 放线菌纲 | 放线菌目 | 微球菌科 | 微杆菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Microbacterium | ||
| 166 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 173 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 177 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 183 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 193 | 放线菌门 | 放线菌纲 | NA | NA | NA | |
| Actinobacteria | Acidimicrobiia | |||||
| 205 | 放线菌门 | 放线菌纲 | 放线菌目 | NA | NA | |
| Actinobacteria | Acidimicrobiia | Acidimicrobiales | ||||
| 207 | 放线菌门 | 嗜热油菌纲 | 索利红杆菌目 | NA | NA | |
| Actinobacteria | Thermoleophilia | Solirubrobacterales | ||||
| 212 | 厚壁菌门 | 芽孢杆菌纲 | 乳杆菌目 | NA | NA | |
| Firmicutes | Bacilli | Lactobacillales | ||||
| 218 | 变形菌门 | β变形菌纲 | NA | NA | NA | |
| Proteobacteria | Betaproteobacteria | |||||
| 233 | 放线菌门 | 放线菌纲 | 放线菌目 | 诺卡氏菌科 | NA | |
| Actinobacteria | Actinobacteria | Actinomycetales | Nocardiaceae | |||
| 253 | 变形菌门 | α变形菌纲 | 红螺菌目 | 红螺菌科 | NA | |
| Proteobacteria | Alphaproteobacteria | Rhodospirillales | Rhodospirillaceae | |||
| 255 | 变形菌门 | β变形菌纲 | NA | NA | NA | |
| Proteobacteria | Betaproteobacteria | |||||
| 271 | 放线菌门 | 放线菌纲 | 放线菌目 | 动孢囊菌科 | 动球菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Kineosporiaceae | Kineococcus | ||
| 274 | 放线菌门 | 放线菌纲 | 放线菌目 | 微球菌科 | 寒冷杆菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Frigoribacterium | ||
| 278 | 放线菌门 | 放线菌纲 | 放线菌目 | 诺卡氏菌科 | 马红球菌 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Nocardiaceae | Rhodococcus | ||
| 301 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | Kaistobacter | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | |||
| 303 | 变形菌门 | β变形菌纲 | NA | NA | NA | |
| Proteobacteria | Betaproteobacteria | |||||
| 313 | 变形菌门 | γ变形菌纲 | 假单胞菌目 | 假单胞菌科 | NA | |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | |||
| 323 | 放线菌门 | 放线菌纲 | 放线菌目 | 动孢囊菌科 | 动球菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Kineosporiaceae | Kineococcus | ||
| 326 | 放线菌门 | 放线菌纲 | 放线菌目 | 微杆菌科 | 寒冷杆菌属 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Frigoribacterium | ||
| 354 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | Kaistobacter | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | |||
| 359 | 变形菌门 | β变形菌纲 | NA | NA | NA | |
| Proteobacteria | Betaproteobacteria | |||||
| 369 | 变形菌门 | γ变形菌纲 | 假单胞菌目 | 莫拉氏菌属 | 不动杆菌属 | |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | Acinetobacter | ||
| 370 | 变形菌门 | γ变形菌纲 | 假单胞菌目 | 假单胞菌科 | NA | |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | |||
| 侵略 | 186 | NA | NA | NA | NA | NA |
| Aggression | 190 | 变形菌门 | NA | NA | NA | NA |
| Proteobacteria | ||||||
| 200 | 变形菌门 | α变形菌纲 | NA | NA | NA | |
| Proteobacteria | Alphaproteobacteria | |||||
| 利他 | 186 | NA | NA | NA | NA | NA |
| Altruism | 190 | 变形菌门 | NA | NA | NA | NA |
| Proteobacteria | ||||||
| 200 | 变形菌门 | α变形菌纲 | NA | NA | NA | |
| Proteobacteria | Alphaproteobacteria | |||||
| 217 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | NA | NA | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | ||||
| 254 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | NA | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | |||
| 302 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 355 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | ||
| 214 | 变形菌门 | α变形菌纲 | 根瘤菌目 | NA | NA | |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | ||||
| 249 | 变形菌门 | α变形菌纲 | 根瘤菌目 | 甲基杆菌科 | NA | |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | |||
| 297 | 变形菌门 | α变形菌纲 | 根瘤菌目 | 甲基杆菌科 | 甲基杆菌属 | |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | Methylobacterium | ||
| 55 | 变形菌门 | α变形菌纲 | 根瘤菌目 | 甲基杆菌科 | 甲基杆菌属 | |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | Methylobacterium | ||
| 33 | 变形菌门 | α变形菌纲 | 鞘脂单胞菌目 | 鞘氨醇单胞菌科 | 鞘氨醇单胞菌属 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas |
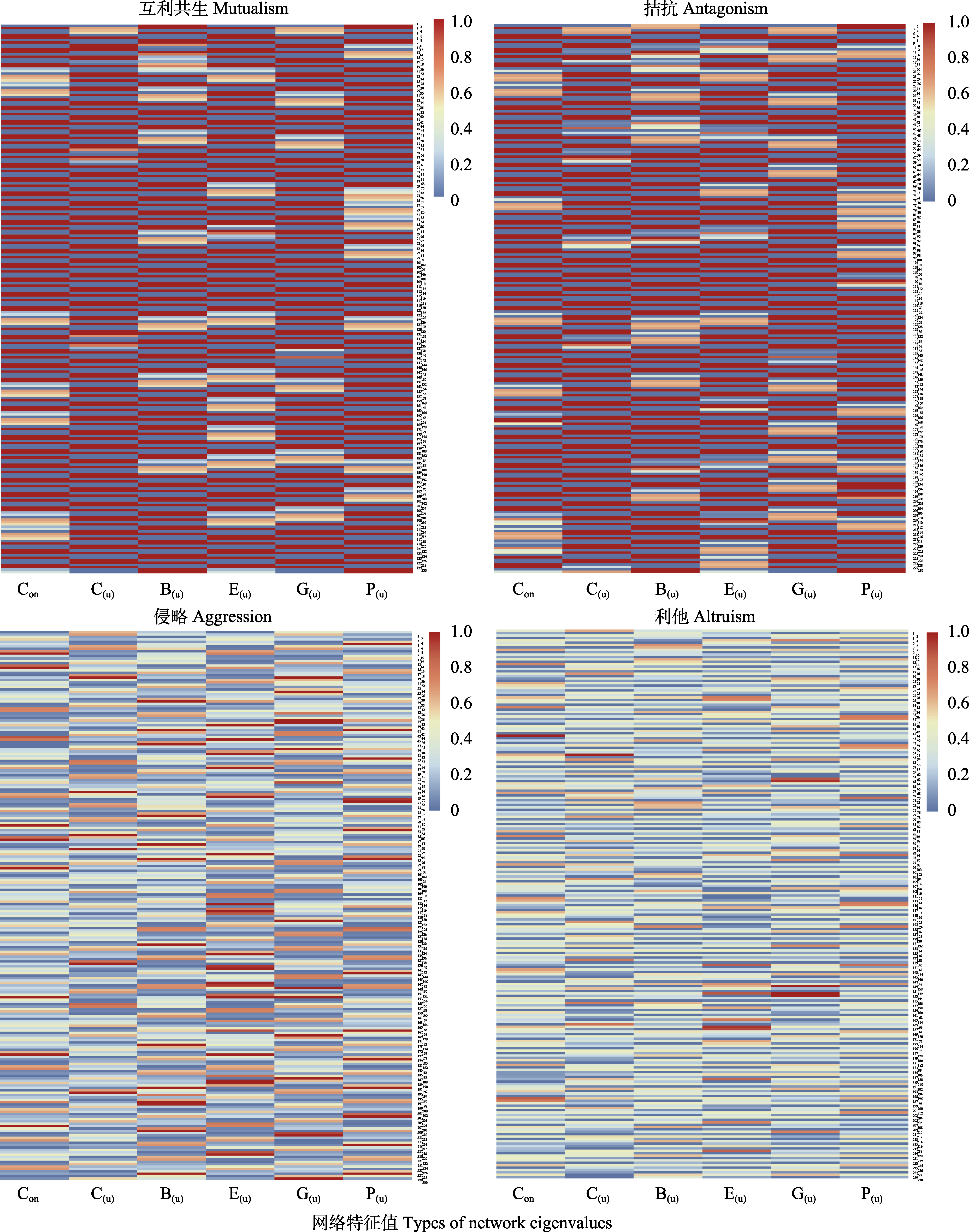
图2 玉米叶际微生物组4个共现网络的6种网络特征值。横坐标从左到右依次代表6种网络特征值, 纵坐标代表230个不同基因型玉米样本, 展现不同个体之间、不同共现网络之间网络特征值的差异。图右边0-1.0的颜色变化代表差异的大小, 0代表没有差异, 1.0代表差异最大。六种网络特征值分别是: 连通性(Con)、接近中心度(C(u))、中介中心度(B(u))、偏心率(E(u))、特征向量(G(u))、网页排名(P(u))。
Fig. 2 Six network indices of four co-occurrence networks of maize-phyllospheric microbiome. As shown in the figures above, the abscissa represents six network feature values from left to right, and the ordinate represents 230 different genotype maize samples, showing the difference in network feature values between different individuals and between different co-occurring networks. The color change of 0-1.0 on the right side of the figure represents the size of the difference, 0 represents no difference, and 1.0 represents the largest difference. The six types of network eigenvalues are: connectivity (Con), closeness (C(u)), betweenness (B(u)), eccentricity (E(u)), eigenvector (G(u)) and pagerank (P(u)).
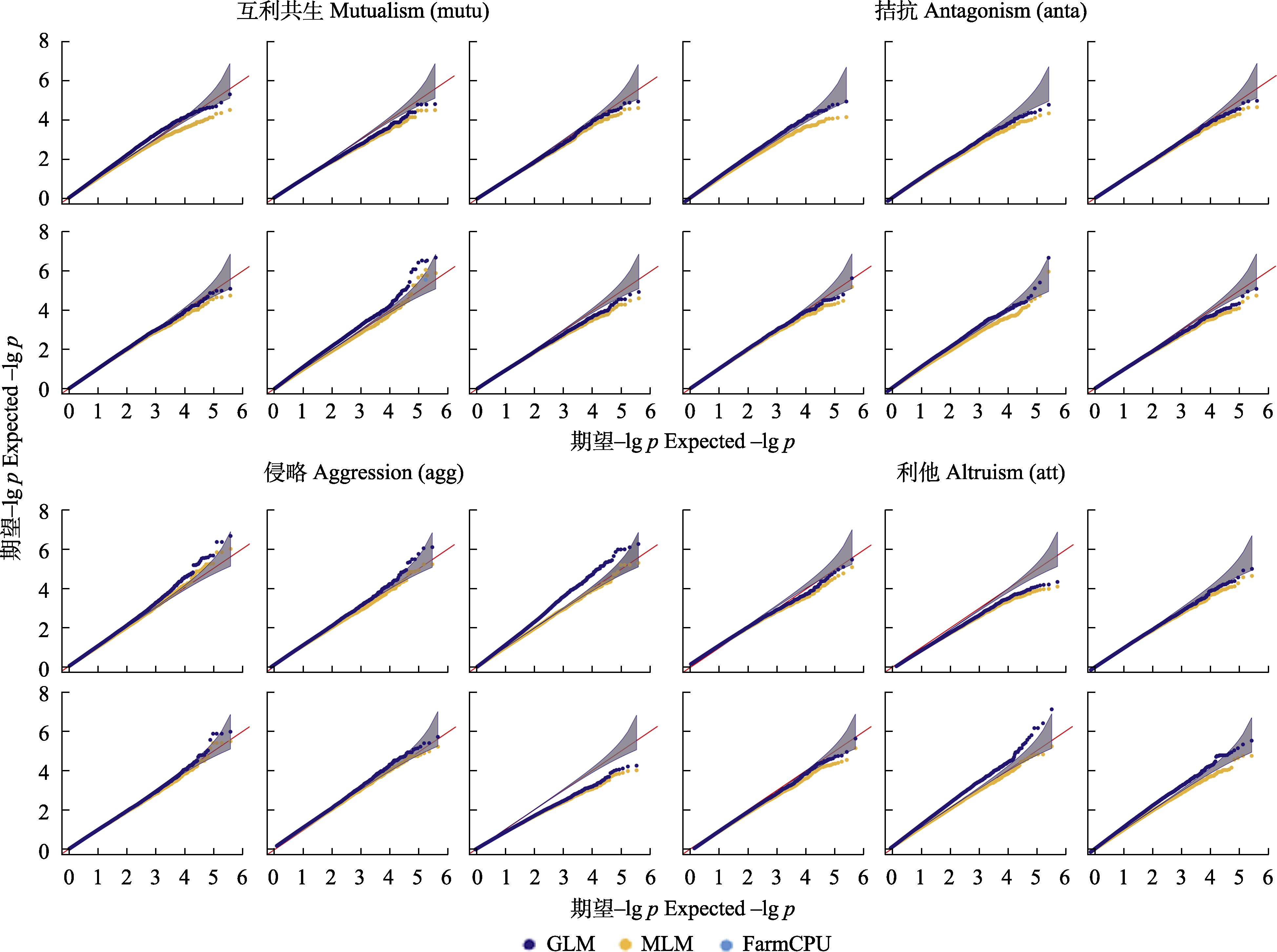
图3 四种相互作用关系下广义线性模型(GLM)、混合线性模型(MLM)、FarmCPU的比较。通过网络作图得到GLM、MLM、FarmCPU模型并用分位图进行展示。四种互作关系下每个网络特征值进行3种模型的比较, 最接近中间红色线的为最优模型。
Fig. 3 Comparison of General Linear Model (GLM), Mixed Linear Model (MLM), Fixed and Random Model Circulating Probability Unification (FarmCPU) under four interaction relationships. GLM, MLM, and FarmCPU models are obtained through network mapping and displayed with Quantile-Quantile diagrams. The indices of each network under the four interaction relationships are compared by three models, and the one closest to the middle red line is the optimal model.
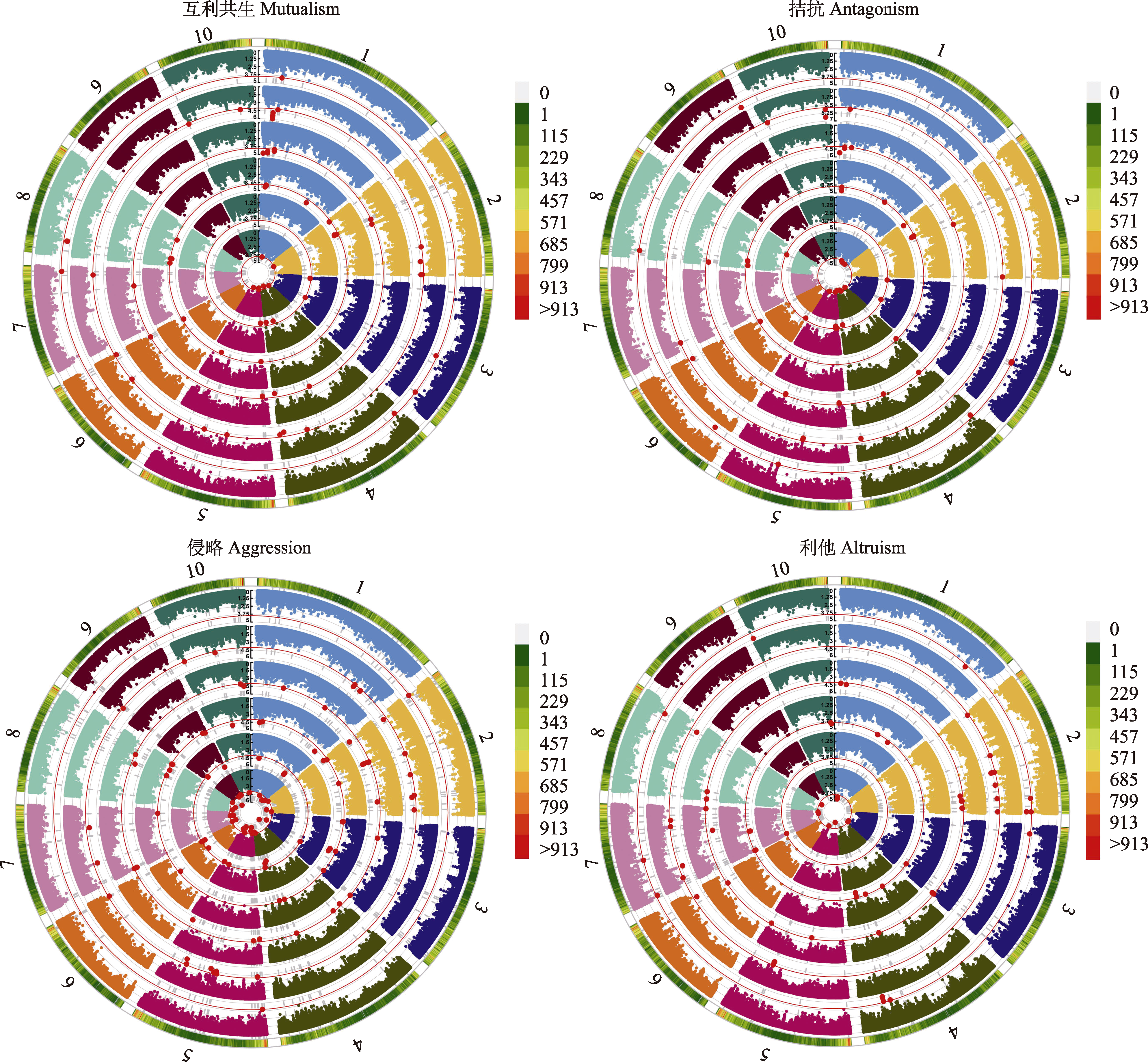
图4 影响玉米叶际微生物的显著单核苷酸多样性(SNPs)位点。基于每种关系中每个网络特征值的最优模型进行可视化, 外圆圈的数字代表的是染色体编号, 内层的6个圆圈从里到外依次是连通性(Con)、接近中心度(C(u))、中介中心度(B(u))、偏心率(E(u))、特征向量(G(u))、网页排名(P(u))最优模型的关联结果, 并把显著阈值设为-lg p ≥ 4, 红色线为阈值线, 红色的点为4种关系下6个网络特征值的显著SNPs。
Fig. 4 Significant single-nucleotide polymorphisms (SNPs) locus affecting phyllospheric microbiome of maize. Visualization based on the optimal model of each network eigenvalue in each relationship. The number of the outer circle represents the chromosome number, and the outermost circle represents the association result of the optimal model of connectivity (Con), closeness (C(u)), betweenness (B(u)), eccentricity (E(u)), eigenvector (G(u)), and pagerank (P(u)) from the inside to the outside, the significance threshold is set to -lg p ≥ 4, the red line is the threshold line, and the red point is the significant SNPs locus for the six network feature values under the four relationships.
| 染色体 Chromosome | 位点 Position | p | 基因编号 Gene_ID | 基因注释 Gene model description |
|---|---|---|---|---|
| chr8_80293025 | 79811419 | 1.22E-05 | LOC103637041 | 染色质修饰相关蛋白EAF1B Chromatin modification-related protein EAF1B |
| chr5_182759726 | 182810285 | 2.35E-05 | LOC100382616 | 转导蛋白/WD40重复序列超家族蛋白 Transducin/WD40 repeat-like superfamily protein |
| chr7_111885528 | 111913781 | 8.54E-05 | LOC100280930 | 乳草pod1基因 Milkweed pod1 |
| chr1_43064775 | 43069297 | 3.85E-05 | LOC100281305 | CDP-甘油二酯-甘油-3-磷酸-3-磷脂酰基转移酶 CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase |
| chr9_22101563 | 22112939 | 5.27E-05 | LOC103639798 | 未鉴定的蛋白质LOC103639798 Uncharacterized protein LOC103639798 |
| chr6_161132530 | 161328052 | 6.32E-05 | LOC103631541 | 尼卡斯特林蛋白 Nicastrin |
| chr9_22101536 | 22112912 | 6.87E-05 | LOC103639798 | 未鉴定的蛋白质LOC103639798 Uncharacterized protein LOC103639798 |
| chr6_161132399 | 161327921 | 9.60E-05 | LOC103631541 | 尼卡斯特林蛋白 Nicastrin |
| chr2_202641758 | 203297345 | 3.40E-05 | LOC103647630 | 磷脂酶D Phospholipase D |
| chr5_18702776 | 18724157 | 6.27E-05 | LOC109939639 | 类枯草杆菌蛋白酶 Subtilisin-like protease |
| chr3_225340724 | 225426794 | 9.21E-05 | LOC100273771 | DENN结构域与WD重复蛋白SCD1 DENN domain and WD repeat-containing protein SCD1 |
| chr5_11847610 | 11859107 | 6.62E-05 | LOC103625953 | 磷脂酰肌醇4-激酶α1 Phosphatidylinositol 4-kinase α1 |
| chr7_9366020 | 9371186 | 8.52E-05 | LOC100381993 | 未鉴定的蛋白LOC100381993 Uncharacterized LOC100381993 |
| chr3_202433096 | 202505131 | 9.00E-05 | LOC100383988 | 含五肽重复序列的At2g27800 Pentatricopeptide repeat-containing protein At2g27800 |
| chr2_203522337 | 204177924 | 4.25E-05 | LOC109944383 | 未鉴定的蛋白LOC109944383 Uncharacterized LOC109944383 |
| chr2_108870299 | 109500380 | 9.71E-05 | LOC103646815 | 未鉴定的蛋白LOC103646815 Uncharacterized LOC103646815 |
| chr2_202739633 | 203395220 | 8.06E-05 | LOC100382522 | 含ACT结构域的蛋白激酶家族蛋白 Putative ACT-domain containing protein kinase family protein |
| chr5_1568127 | 1568410 | 9.25E-05 | LOC732836 | 可能的离子通道CASTOR Probable ion channel CASTOR |
| chr2_175336727 | 175982640 | 8.94E-05 | LOC103647325 | 丝氨酸/苏氨酸蛋白磷酸酶2A57kDa调节亚基B′ Serine/threonine protein phosphatase 2A57kDa regulatory subunit B′ |
| chr4_167034562 | 167064438 | 5.87E-05 | LOC103653988 | 未鉴定的蛋白LOC103653988 Uncharacterized LOC103653988 |
| chr4_11075888 | 11082944 | 6.84E-05 | LOC103652818 | 受体样蛋白激酶HSL1 Receptor-like protein kinase HSL1 |
| chr8_5531988 | 5532528 | 3.40E-05 | LOC103634691 | 未鉴定的蛋白LOC103634691 Uncharacterized LOC103634691 |
| chr5_95630987 | 95668349 | 5.36E-05 | LOC100272320 | 蛋白酶体β亚基 Proteasome subunit beta type |
表2 影响玉米叶际微生物网络的枢纽基因
Table 2 Hub genes that affect maize leaf microbiome networks
| 染色体 Chromosome | 位点 Position | p | 基因编号 Gene_ID | 基因注释 Gene model description |
|---|---|---|---|---|
| chr8_80293025 | 79811419 | 1.22E-05 | LOC103637041 | 染色质修饰相关蛋白EAF1B Chromatin modification-related protein EAF1B |
| chr5_182759726 | 182810285 | 2.35E-05 | LOC100382616 | 转导蛋白/WD40重复序列超家族蛋白 Transducin/WD40 repeat-like superfamily protein |
| chr7_111885528 | 111913781 | 8.54E-05 | LOC100280930 | 乳草pod1基因 Milkweed pod1 |
| chr1_43064775 | 43069297 | 3.85E-05 | LOC100281305 | CDP-甘油二酯-甘油-3-磷酸-3-磷脂酰基转移酶 CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase |
| chr9_22101563 | 22112939 | 5.27E-05 | LOC103639798 | 未鉴定的蛋白质LOC103639798 Uncharacterized protein LOC103639798 |
| chr6_161132530 | 161328052 | 6.32E-05 | LOC103631541 | 尼卡斯特林蛋白 Nicastrin |
| chr9_22101536 | 22112912 | 6.87E-05 | LOC103639798 | 未鉴定的蛋白质LOC103639798 Uncharacterized protein LOC103639798 |
| chr6_161132399 | 161327921 | 9.60E-05 | LOC103631541 | 尼卡斯特林蛋白 Nicastrin |
| chr2_202641758 | 203297345 | 3.40E-05 | LOC103647630 | 磷脂酶D Phospholipase D |
| chr5_18702776 | 18724157 | 6.27E-05 | LOC109939639 | 类枯草杆菌蛋白酶 Subtilisin-like protease |
| chr3_225340724 | 225426794 | 9.21E-05 | LOC100273771 | DENN结构域与WD重复蛋白SCD1 DENN domain and WD repeat-containing protein SCD1 |
| chr5_11847610 | 11859107 | 6.62E-05 | LOC103625953 | 磷脂酰肌醇4-激酶α1 Phosphatidylinositol 4-kinase α1 |
| chr7_9366020 | 9371186 | 8.52E-05 | LOC100381993 | 未鉴定的蛋白LOC100381993 Uncharacterized LOC100381993 |
| chr3_202433096 | 202505131 | 9.00E-05 | LOC100383988 | 含五肽重复序列的At2g27800 Pentatricopeptide repeat-containing protein At2g27800 |
| chr2_203522337 | 204177924 | 4.25E-05 | LOC109944383 | 未鉴定的蛋白LOC109944383 Uncharacterized LOC109944383 |
| chr2_108870299 | 109500380 | 9.71E-05 | LOC103646815 | 未鉴定的蛋白LOC103646815 Uncharacterized LOC103646815 |
| chr2_202739633 | 203395220 | 8.06E-05 | LOC100382522 | 含ACT结构域的蛋白激酶家族蛋白 Putative ACT-domain containing protein kinase family protein |
| chr5_1568127 | 1568410 | 9.25E-05 | LOC732836 | 可能的离子通道CASTOR Probable ion channel CASTOR |
| chr2_175336727 | 175982640 | 8.94E-05 | LOC103647325 | 丝氨酸/苏氨酸蛋白磷酸酶2A57kDa调节亚基B′ Serine/threonine protein phosphatase 2A57kDa regulatory subunit B′ |
| chr4_167034562 | 167064438 | 5.87E-05 | LOC103653988 | 未鉴定的蛋白LOC103653988 Uncharacterized LOC103653988 |
| chr4_11075888 | 11082944 | 6.84E-05 | LOC103652818 | 受体样蛋白激酶HSL1 Receptor-like protein kinase HSL1 |
| chr8_5531988 | 5532528 | 3.40E-05 | LOC103634691 | 未鉴定的蛋白LOC103634691 Uncharacterized LOC103634691 |
| chr5_95630987 | 95668349 | 5.36E-05 | LOC100272320 | 蛋白酶体β亚基 Proteasome subunit beta type |
| [1] |
Asaf S, Numan M, Khan AL, Al-Harrasi A (2020). Sphingomonas: from diversity and genomics to functional role in environmental remediation and plant growth. Critical Reviews in Biotechnology, 40, 138-152.
DOI PMID |
| [2] |
Barkan A, Small I (2014). Pentatricopeptide repeat proteins in plants. Annual Review of Plant Biology, 65, 415-442.
DOI PMID |
| [3] |
Batstone Rebecca T, O’brien Anna M, Harrison Tia L, Frederickson Megan E (2020). Experimental evolution makes microbes more cooperative with their local host genotype. Science, 370, 476-478.
DOI PMID |
| [4] |
Beilsmith K, Thoen MPM, Brachi B, Gloss AD, Khan MH, Bergelson J (2019). Genome-wide association studies on the phyllosphere microbiome: embracing complexity in host-microbe interactions. The Plant Journal, 97, 164-181.
DOI PMID |
| [5] | Bergelson J, Mittelstrass J, Horton MW (2019). Characterizing both bacteria and fungi improves understanding of the Arabidopsis root microbiome. Scientific Reports, 9, 24. DOI: 10.1038/s41598-018-37208-z. |
| [6] | Brachi B, Filiault D, Darme P, Mentec M, Kerdaffrec E, Rabanal F, Anastasio A, Box M, Duncan S, Morton T, Novikova P, Perisin M, Tsuchimatsu T, Woolley R, Yu M, et al. (2017). Plant genes influence microbial hubs that shape beneficial leaf communities. bioRxiv. DOI: 10.1101/181198. |
| [7] | Brachi B, Filiault D, Whitehurst H, Darme P, Le Gars P, Le Mentec M, Morton TC, Kerdaffrec E, Rabanal F, Anastasio A, Box MS, Duncan S, Huang F, Leff R, Novikova P, et al. (2022). Plant genetic effects on microbial hubs impact host fitness in repeated field trials. Proceedings of the National Academy of Sciences of the United States of America, 119, e2201285119. DOI: 10.1073/pnas.2201285119. |
| [8] |
Carrión VJ, Perez-Jaramillo J, Cordovez V, Tracanna V, de Hollander M, Ruiz-Buck D, Mendes LW, Gomez-Exposito R, Elsayed SS, Mohanraju P, Arifah A, van der Oost J, Paulson JN, Mendes R, et al. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science, 366, 606-612.
DOI PMID |
| [9] | Chen T, Nomura K, Wang XL, Sohrabi R, Xu J, Yao L, Paasch BC, Ma L, Kremer J, Cheng Y, Zhang L, Wang N, Wang E, Xin X, He S (2020). A plant genetic network for preventing dysbiosis in the phyllosphere. Nature, 580, 653-657. |
| [10] | de Vries FT, Griffiths RI, Knight CG, Nicolitch O, Williams A (2020). Harnessing rhizosphere microbiomes for drought- resilient crop production. Science, 368, 270-274. |
| [11] | Delaux P, Schornack S (2021). Plant evolution driven by interactions with symbiotic and pathogenic microbes. Science, 371, eaba6605. DOI: 10.1126/science.aba6605. |
| [12] | Deng S, Caddell DF, Xu G, Dahlen L, Washington L, Yang J, Coleman-Derr D (2021). Genome wide association study reveals plant loci controlling heritability of the rhizosphere microbiome. The ISME Journal, 15, 3181-3194. |
| [13] |
Distéfano AM, Scuffi D, García-Mata C, Lamattina L, Laxalt AM (2012). Phospholipase δD is involved in nitric oxide-induced stomatal closure. Planta, 236, 1899-1907.
DOI PMID |
| [14] | Escudero-Martinez C, Coulter M, Alegria Terrazas R, Foito A, Kapadia R, Pietrangelo L, Maver M, Sharma R, Aprile A, Morris J, Hedley PE, Maurer A, Pillen K, Naclerio G, Mimmo T, et al. (2022). Identifying plant genes shaping microbiota composition in the barley rhizosphere. Nature Communications, 13, 3443. DOI: 10.1038/s41467-022-31022-y. |
| [15] |
Falbel TG, Koch LM, Nadeau JA, Segui-Simarro JM, Sack FD, Bednarek SY (2003). SCD1 is required for cell cytokinesis and polarized cell expansion in Arabidopsis thaliana. Development, 130, 4011-4024.
PMID |
| [16] |
Figueiredo J, Sousa Silva M, Figueiredo A (2018). Subtilisin- like proteases in plant defence: the past, the present and beyond. Molecular Plant Pathology, 19, 1017-1028.
DOI PMID |
| [17] |
Finkel OM, Castrillo G, Herrera Paredes S, Salas González I, Dangl JL (2017). Understanding and exploiting plant beneficial microbes. Current Opinion in Plant Biology, 38, 155-163.
DOI PMID |
| [18] |
Gong TY, Xin XF (2021). Phyllosphere microbiota: community dynamics and its interaction with plant hosts. Journal of Integrative Plant Biology, 63, 297-304.
DOI |
| [19] |
Hawkes CV, Kjøller R, Raaijmakers JM, Riber L, Christensen S, Rasmussen S, Christensen JH, Dahl AB, Westergaard JC, Nielsen M, Brown-Guedira G, Hestbjerg Hansen L (2021). Extension of plant phenotypes by the foliar microbiome. Annual Review of Plant Biology, 72, 823-846.
DOI PMID |
| [20] | He XQ, Zhang Q, Li BB, Jin Y, Jiang LB, Wu RL (2021). Network mapping of root-microbe interactions in Arabidopsis thaliana. NPJ Biofilms and Microbiomes, 7, 72. DOI: 10.1038/s41522-021-00241-4. |
| [21] | Horton MW, Bodenhausen N, Beilsmith K, Meng DZ, Muegge BD, Subramanian S, Vetter MM, Vilhjálmsson BJ, Nordborg M, Gordon JI, Bergelson J (2014). Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nature Communications, 5, 5320. DOI: 10.1038/ncomms6320. |
| [22] | Jiang L, Liu X, He X, Jin Y, Cao Y, Zhan X, Griffin CH, Gragnoli C, Wu R (2021). A behavioral model for mapping the genetic architecture of gut-microbiota networks. Gut Microbes, 13, 1820847. DOI: 10.1080/19490976.2020.1820847. |
| [23] | Li KH, Cheng KX, Wang HC, Zhang Q, Yang Y, Jin Y, He XQ, Wu RL (2022a). Disentangling leaf-microbiome interactions in Arabidopsis thaliana by network mapping. Frontiers in Plant Science, 13, 996121. DOI: 10.3389/fpls.2022.996121. |
| [24] | Li KH, Wang HC, Cheng KX, Yang Y, Jin Y, He XQ (2023). Genetic mechanisms of plant-microbiome interaction by genome-wide association analysis study. Biotechnology Bulletin, 39(2), 24-34. |
|
[李凯航, 王浩臣, 程可心, 杨艳, 金一, 何晓青 (2023). 全基因组关联分析研究植物与微生物组的互作遗传机制. 生物技术通报, 39(2), 24-34.]
DOI |
|
| [25] | Li PD, Zhu ZR, Zhang YZ, Xu JP, Wang HK, Wang ZY, Li HY (2022b). The phyllosphere microbiome shifts toward combating melanose pathogen. Microbiome, 10, 56. DOI: 10.1186/s40168-022-01234-x. |
| [26] | Li Z, Bai X, Jiao S, Li Y, Li P, Yang Y, Zhang H, Wei G (2021). A simplified synthetic community rescues Astragalus mongholicus from root rot disease by activating plant- induced systemic resistance. Microbiome, 9, 217. DOI: 10.1186/s40168-021-01169-9. |
| [27] | Madhaiyan M, Alex THH, Te Ngoh S, Prithiviraj B, Ji L (2015). Leaf-residing Methylobacterium species fix nitrogen and promote biomass and seed production in Jatropha curcas. Biotechnology for Biofuels, 8, 1-14. |
| [28] | Meier MA, Xu G, Lopez-Guerrero MG, Li G, Smith C, Sigmon B, Herr JR, Alfano JR, Ge Y, Schnable JC, Yang J (2022). Association analyses of host genetics, root- colonizing microbes, and plant phenotypes under different nitrogen conditions in maize. eLife, 11, 75790. DOI: 10.7554/eLife.75790. |
| [29] | Müller DB, Vogel C, Bai Y, Vorholt JA (2016). The plant microbiota: systems-level insights and perspectives. Annual Review of Genetics, 50, 211-234. |
| [30] | Oyserman BO, Flores SS, Griffioen T, Pan X, van der Wijk E, Pronk L, Lokhorst W, Nurfikari A, Paulson JN, Movassagh M, Stopnisek N, Kupczok A, Cordovez V, Carrión VJ, Ligterink W, et al. (2022). Disentangling the genetic basis of rhizosphere microbiome assembly in tomato. Nature Communications, 13, 3228. DOI: 10.1038/s41467-022-30849-9. |
| [31] | Pang Z, Chen J, Wang T, Gao C, Li Z, Guo L, Xu J, Cheng Y (2021). Linking plant secondary metabolites and plant microbiomes: a review. Frontiers in Plant Science, 12, 621276. DOI: 10.3389/fpls.2021.621276. |
| [32] | Roman-Reyna V, Pinili D, Borja FN, Quibod IL, Groen SC, Alexandrov N, Mauleon R, Oliva R (2020). Characterization of the leaf microbiome from whole-genome sequencing data of the 3000 rice genomes project. Rice, 13, 72. DOI: 10.1186/s12284-020-00432-1. |
| [33] |
Schlaeppi K, Bulgarelli D (2015). The plant microbiome at work. Molecular Plant-Microbe Interactions, 28, 212-217.
DOI PMID |
| [34] | Schmitz L, Yan Z, Schneijderberg M, de Roij M, Pijnenburg R, Zheng Q, Franken C, Dechesne A, Trindade LM, van Velzen R, Bisseling T, Geurts R, Cheng X (2022). Synthetic bacterial community derived from a desert rhizosphere confers salt stress resilience to tomato in the presence of a soil microbiome. The ISME Journal, 16, 1907-1920. |
| [35] | Stringlis IA, Yu K, Feussner K, de Jonge R, van Bentum S, van Verk MC, Berendsen RL, Bakker PAHM, Feussner I, Pieterse CMJ (2018). MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proceedings of the National Academy of Sciences of the United States of America, 115, E5213-E5222. |
| [36] |
Tan XF, Xie HT, Yu JW, Wang YF, Xu JM, Xu P, Ma B (2022). Host genetic determinants drive compartment-specific assembly of tea plant microbiomes. Plant Biotechnology Journal, 20, 2174-2186.
DOI PMID |
| [37] | Tong H, Zheng C, Li B, Swanner ED, Liu C, Chen M, Xia Y, Liu Y, Ning Z, Li F, Feng X (2021). Microaerophilic oxidation of Fe(II) coupled with simultaneous carbon fixation and As(III) oxidation and sequestration in karstic paddy soil. Environmental Science & Technology, 55, 3634-3644. |
| [38] | Trivedi P, Leach JE, Tringe SG, Sa TM, Singh BK (2020). Plant-microbiome interactions: from community assembly to plant health. Nature Reviews Microbiology, 18, 607-621. |
| [39] | Vogel CM, Potthoff DB, Schäfer M, Barandun N, Vorholt JA (2021). Protective role of the Arabidopsis leaf microbiota against a bacterial pathogen. Nature Microbiology, 6, 1537-1548. |
| [40] |
Wagner MR, Busby PE, Balint-Kurti P (2020). Analysis of leaf microbiome composition of near-isogenic maize lines differing in broad-spectrum disease resistance. New Phytologist, 225, 2152-2165.
DOI PMID |
| [41] | Wallace JG, Kremling KA, Kovar LL, Buckler ES (2018). Quantitative genetics of the maize leaf microbiome. Phytobiomes Journal, 2, 208-224. |
| [42] |
Wang CH, Li YJ, Li MJ, Zhang KF, Ma WJ, Zheng L, Xu HY, Cui BF, Liu R, Yang YQ, Zhong YJ, Liao H (2021). Functional assembly of root-associated microbial consortia improves nutrient efficiency and yield in soybean. Journal of Integrative Plant Biology, 63, 1021-1035.
DOI |
| [43] | Wang YY, Wang XL, Sun SA, Jin CZ, Su JM, Wei JP, Luo X, Wen JW, Wei T, Sahu SK, Zou HF, Chen HY, Mu ZX, Zhang GY, Liu X, et al. (2022). GWAS, MWAS and mGWAS provide insights into precision agriculture based on genotype- dependent microbial effects in foxtail millet. Nature Communications, 13, 5913. DOI: 10.1038/s41467-022-33238-4. |
| [44] | Wang ZH, Song Y (2022). Toward understanding the genetic bases underlying plant-mediated “cry for help” to the microbiota. iMeta, 1, e8. DOI: 10.1002/imt2.8. |
| [45] | Xie YH, Jia P, Zheng XT, Li JT, Shu WS, Wang YT (2022). Impacts and action pathways of domestication on diversity and community structure of crop microbiome: a review. Chinese Journal of Plant Ecology, 46, 249-266. |
|
[谢育杭, 贾璞, 郑修坛, 李金天, 束文圣, 王宇涛 (2022). 驯化对作物微生物组多样性和群落结构的影响及作用途径. 植物生态学报, 46, 249-266.]
DOI |
|
| [46] | Xiong C, Zhu YG, Wang JT, Singh B, Han LL, Shen JP, Li PP, Wang GB, Wu CF, Ge AH, Zhang LM, He JZ (2021). Host selection shapes crop microbiome assembly and network complexity. New Phytologist, 229, 1091-1104. |
| [47] | Yang K, Wang HL, Ye KH, Wang P, Meng GY, Luo C, Guo LW (2021). Advances in research on phyllosphere microorganisms and their interaction with plants. Journal of Yunnan Agricultural University (Natural Science), 36(1), 155-164. |
| [杨宽, 王慧玲, 叶坤浩, 王佩, 孟广云, 罗成, 郭力维 (2021). 叶际微生物及与植物互作的研究进展. 云南农业大学学报(自然科学), 36(1), 155-164.] | |
| [48] | Yin C, Casa Vargas JM, Schlatter DC, Hagerty CH, Hulbert SH, Paulitz TC (2021a). Rhizosphere community selection reveals bacteria associated with reduced root disease. Microbiome, 9, 86. DOI: 10.1186/s40168-020-00997-5. |
| [49] | Yin LL, Zhang HH, Tang ZS, Xu JY, Yin D, Zhang ZW, Yuan XH, Zhu MJ, Zhao SH, Li XY, Liu X (2021b). rMVP: a memory-efficient, visualization-enhanced, and parallel- accelerated tool for genome-wide association study. Genomics, Proteomics & Bioinformatics, 19, 619-628. |
| [50] |
Zhang J, Liu Y, Guo X, Qin Y, Garrido-Oter R, Schulze-Lefert P, Bai Y (2021). High-throughput cultivation and identification of bacteria from the plant root microbiota. Nature Protocols, 16, 988-1012.
DOI PMID |
| [51] |
Zhang JY, Liu YX, Zhang N, Hu B, Jin T, Xu HR, Qin YA, Yan PX, Zhang XN, Guo XX, Hui J, Cao SY, Wang X, Wang C, Wang H, et al. (2019). NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nature Biotechnology, 37, 676-684.
DOI PMID |
| [52] | Zhang LY, Zhang ML, Huang SY, Li LJ, Gao QA, Wang Y, Zhang SQ, Huang SM, Yuan LA, Wen YC, Liu KL, Yu XC, Li DC, Zhang L, Xu XP, et al. (2022). A highly conserved core bacterial microbiota with nitrogen-fixation capacity inhabits the xylem sap in maize plants. Nature Communications, 13, 3361. DOI: 10.1038/s41467-022-31113-w. |
| [1] | 李义博, 宋贺, 周莉, 许振柱, 周广胜. C4植物玉米的光合-光响应曲线模拟研究[J]. 植物生态学报, 2017, 41(12): 1289-1300. |
| [2] | 赵文赛, 孙永林, 刘西平. 干旱-复水-再干旱处理对玉米光合能力和生长的影响[J]. 植物生态学报, 2016, 40(6): 594-603. |
| [3] | 孟凡超, 张佳华, 姚凤梅. CO2浓度升高和降水增加协同作用对玉米产量及生长发育的影响[J]. 植物生态学报, 2014, 38(10): 1064-1073. |
| [4] | 王艳哲, 邵立威, 刘秀位, 张小雨, 张喜英. 小麦和玉米根系取样位置优化确定及根系分布模拟[J]. 植物生态学报, 2013, 37(4): 365-372. |
| [5] | 李涛, 陈保冬. 丛枝菌根真菌通过上调根系及自身水孔蛋白基因表达提高玉米抗旱性[J]. 植物生态学报, 2012, 36(9): 973-981. |
| [6] | 杨斌, 谢甫绨, 温学发, 孙晓敏, 王建林. 华北平原农田土壤蒸发δ18O的日变化特征及其影响因素[J]. 植物生态学报, 2012, 36(6): 539-549. |
| [7] | 时鹏, 王淑平, 贾书刚, 高强, 孙晓强. 三种种植方式对土壤微生物群落组成的影响[J]. 植物生态学报, 2011, 35(9): 965-972. |
| [8] | 王红丽, 张绪成, 宋尚有. 半干旱区旱地不同覆盖种植方式玉米田的土壤水分和产量效应[J]. 植物生态学报, 2011, 35(8): 825-833. |
| [9] | 冯远娇, 金琼, 王建武. 机械损伤对Bt玉米化学防御的系统诱导效应[J]. 植物生态学报, 2010, 34(6): 695-703. |
| [10] | 贾士芳, 李从锋, 董树亭, 张吉旺. 弱光胁迫影响夏玉米光合效率的生理机制初探[J]. 植物生态学报, 2010, 34(12): 1439-1447. |
| [11] | 冯远娇, 王建武, 骆世明. 叶片涂施茉莉酸对玉米幼苗防御反应的时间和浓度效应[J]. 植物生态学报, 2009, 33(4): 812-823. |
| [12] | 刘天学, 李潮海, 马新明, 赵霞, 刘士英. 不同基因型玉米间作对叶片衰老、籽粒产量和品质的影响[J]. 植物生态学报, 2008, 32(4): 914-921. |
| [13] | 刘广才, 杨祁峰, 李隆, 孙建好. 小麦/玉米间作优势及地上部与地下部因素的相对贡献[J]. 植物生态学报, 2008, 32(2): 477-484. |
| [14] | 陈小莉, 李世清, 任小龙, 强虹, 吉春容, 闫登明. 大气NH3浓度升高对不同氮效率玉米生理指标及生物量的影响[J]. 植物生态学报, 2008, 32(1): 204-211. |
| [15] | 李祎君, 许振柱, 王云龙, 周莉, 周广胜. 玉米农田水热通量动态与能量闭合分析[J]. 植物生态学报, 2007, 31(6): 1132-1144. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
Copyright © 2022 版权所有 《植物生态学报》编辑部
地址: 北京香山南辛村20号, 邮编: 100093
Tel.: 010-62836134, 62836138; Fax: 010-82599431; E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
备案号: 京ICP备16067583号-19