

Chin J Plant Ecol ›› 2018, Vol. 42 ›› Issue (12): 1200-1210.DOI: 10.17521/cjpe.2018.0120
• Research Articles • Previous Articles Next Articles
LU Ying,LI Kun,NI Rui-Qiang,LIANG Qiang,LI Chuan-Rong,ZHANG Cai-Hong( )
)
Received:2018-02-21
Revised:2018-10-31
Online:2018-12-20
Published:2019-04-04
Contact:
Cai-Hong ZHANG
Supported by:LU Ying, LI Kun, NI Rui-Qiang, LIANG Qiang, LI Chuan-Rong, ZHANG Cai-Hong. Effects of fine root decomposition on bacterial community structure of four dominated tree species in Mount Taishan, China[J]. Chin J Plant Ecol, 2018, 42(12): 1200-1210.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.plant-ecology.com/EN/10.17521/cjpe.2018.0120
| 树种 Species | C (%) | N (%) | P (%) | C:N | N:P | 木质素 Lignin (%) |
|---|---|---|---|---|---|---|
| RP | 48.77 ± 0.33c | 3.36 ± 0.002a | 0.53 ± 0.05a | 14.51 ± 0.09d | 6.36 ± 0.34a | 29.59 ± 0.47c |
| QA | 46.39 ± 0.17d | 1.08 ± 0.008b | 0.46 ± 0.01a | 43.02 ± 0.17c | 2.34 ± 0.05b | 33.78 ± 0.60b |
| PD | 54.65 ± 0.17a | 0.38 ± 0.009d | 0.39 ± 0.03b | 142.48 ± 3.72a | 1.00 ± 0.07c | 38.34 ± 0.30a |
| PT | 49.96 ± 0.13b | 0.85 ± 0.004c | 0.41 ± 0.03b | 59.04 ± 0.19b | 2.10 ± 0.14b | 37.78 ± 0.15a |
Table 1 Differences in initial element contents of fine root litter (mean ± SE, n = 3)
| 树种 Species | C (%) | N (%) | P (%) | C:N | N:P | 木质素 Lignin (%) |
|---|---|---|---|---|---|---|
| RP | 48.77 ± 0.33c | 3.36 ± 0.002a | 0.53 ± 0.05a | 14.51 ± 0.09d | 6.36 ± 0.34a | 29.59 ± 0.47c |
| QA | 46.39 ± 0.17d | 1.08 ± 0.008b | 0.46 ± 0.01a | 43.02 ± 0.17c | 2.34 ± 0.05b | 33.78 ± 0.60b |
| PD | 54.65 ± 0.17a | 0.38 ± 0.009d | 0.39 ± 0.03b | 142.48 ± 3.72a | 1.00 ± 0.07c | 38.34 ± 0.30a |
| PT | 49.96 ± 0.13b | 0.85 ± 0.004c | 0.41 ± 0.03b | 59.04 ± 0.19b | 2.10 ± 0.14b | 37.78 ± 0.15a |
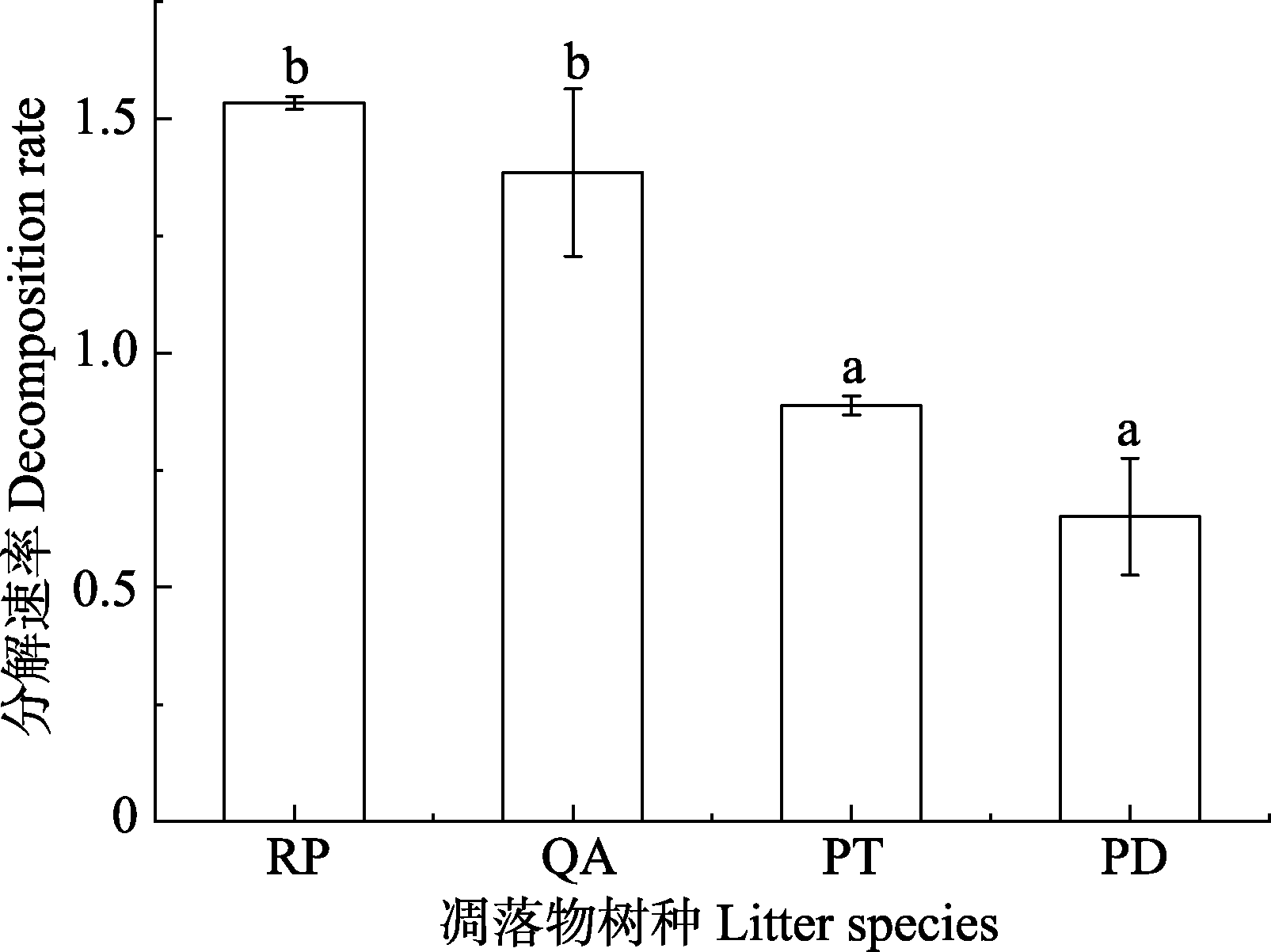
Fig. 1 Difference in decomposition rate among four litter species (mean ± SE) in Mount Taishan. PD, Pinus densiflora; PT, Pinus tabulaeformis; QA, Quercus acutissima; RP, Robinia pseudoacacia. Different lowercase letters represent significant differences among different species (p < 0.05).
| 树种 Species | 物种数 NO. of observed species | 覆盖率Coverage (%) | Chao1指数 Chao1 index | Ace指数 Ace index | 系统发育多样性Phylogenetic diversity | Shannon-Wiener指数Shannon-Wiener index |
|---|---|---|---|---|---|---|
| RP | 2 149 ± 71a | 98.6 ± 0.1b | 3 088.0 ± 140.4ab | 3 062.2 ± 143.5ab | 159.2 ± 4.2a | 8.38 ± 0.59a |
| QA | 1 970 ± 120a | 97.7 ± 0.2a | 2 824.2 ± 88.5a | 2 843.8 ± 62.0a | 147.8 ± 7.6a | 8.14 ± 0.16a |
| PD | 2 759 ± 25b | 98.3 ± 0.2ab | 3 544.7 ± 50.3c | 3 530.6 ± 34.3c | 198.6 ± 5.1b | 8.81 ± 0.35b |
| PT | 2 568 ± 39b | 97.6 ± 0.2a | 3 395.0 ± 2.2bc | 3 341.9 ± 68.4bc | 193.1 ± 3.2b | 8.88 ± 0.18b |
Table 2 Statistical analysis of bacterial diversity in Mount Taishan after one year of fine root decomposition (mean ± SE, n = 3)
| 树种 Species | 物种数 NO. of observed species | 覆盖率Coverage (%) | Chao1指数 Chao1 index | Ace指数 Ace index | 系统发育多样性Phylogenetic diversity | Shannon-Wiener指数Shannon-Wiener index |
|---|---|---|---|---|---|---|
| RP | 2 149 ± 71a | 98.6 ± 0.1b | 3 088.0 ± 140.4ab | 3 062.2 ± 143.5ab | 159.2 ± 4.2a | 8.38 ± 0.59a |
| QA | 1 970 ± 120a | 97.7 ± 0.2a | 2 824.2 ± 88.5a | 2 843.8 ± 62.0a | 147.8 ± 7.6a | 8.14 ± 0.16a |
| PD | 2 759 ± 25b | 98.3 ± 0.2ab | 3 544.7 ± 50.3c | 3 530.6 ± 34.3c | 198.6 ± 5.1b | 8.81 ± 0.35b |
| PT | 2 568 ± 39b | 97.6 ± 0.2a | 3 395.0 ± 2.2bc | 3 341.9 ± 68.4bc | 193.1 ± 3.2b | 8.88 ± 0.18b |
| C (%) | N (%) | P (%) | C:N | N:P | 木质素 Lignin (%) | |
|---|---|---|---|---|---|---|
| 物种数 NO. Of observed species | 0.884** | -0.541 | 0.679* | 0.790* | -0.496 | 0.726* |
| 覆盖率 Coverage (%) | 0.331 | 0.437 | -0.482 | 0.126 | 0.517 | -0.344 |
| Chao1指数 Chao1 index | 0.858** | -0.413 | 0.608 | 0.706* | -0.377 | 0.642 |
| Ace指数 Ace index | 0.874** | -0.446 | 0.593 | 0.748* | -0.405 | 0.661* |
| 系统发育多样性 Phylogenetic diversity | 0.829* | -0.547 | 0.744* | 0.730* | -0.515 | 0.749* |
| Shannon-Wiener指数 Shannon-Wiener index | 0.552 | -0.292 | 0.491 | 0.378 | -0.246 | 0.418 |
Table 3 Correlation analysis between bacterial α diversity and the initial properties of litter after one year of decomposition
| C (%) | N (%) | P (%) | C:N | N:P | 木质素 Lignin (%) | |
|---|---|---|---|---|---|---|
| 物种数 NO. Of observed species | 0.884** | -0.541 | 0.679* | 0.790* | -0.496 | 0.726* |
| 覆盖率 Coverage (%) | 0.331 | 0.437 | -0.482 | 0.126 | 0.517 | -0.344 |
| Chao1指数 Chao1 index | 0.858** | -0.413 | 0.608 | 0.706* | -0.377 | 0.642 |
| Ace指数 Ace index | 0.874** | -0.446 | 0.593 | 0.748* | -0.405 | 0.661* |
| 系统发育多样性 Phylogenetic diversity | 0.829* | -0.547 | 0.744* | 0.730* | -0.515 | 0.749* |
| Shannon-Wiener指数 Shannon-Wiener index | 0.552 | -0.292 | 0.491 | 0.378 | -0.246 | 0.418 |
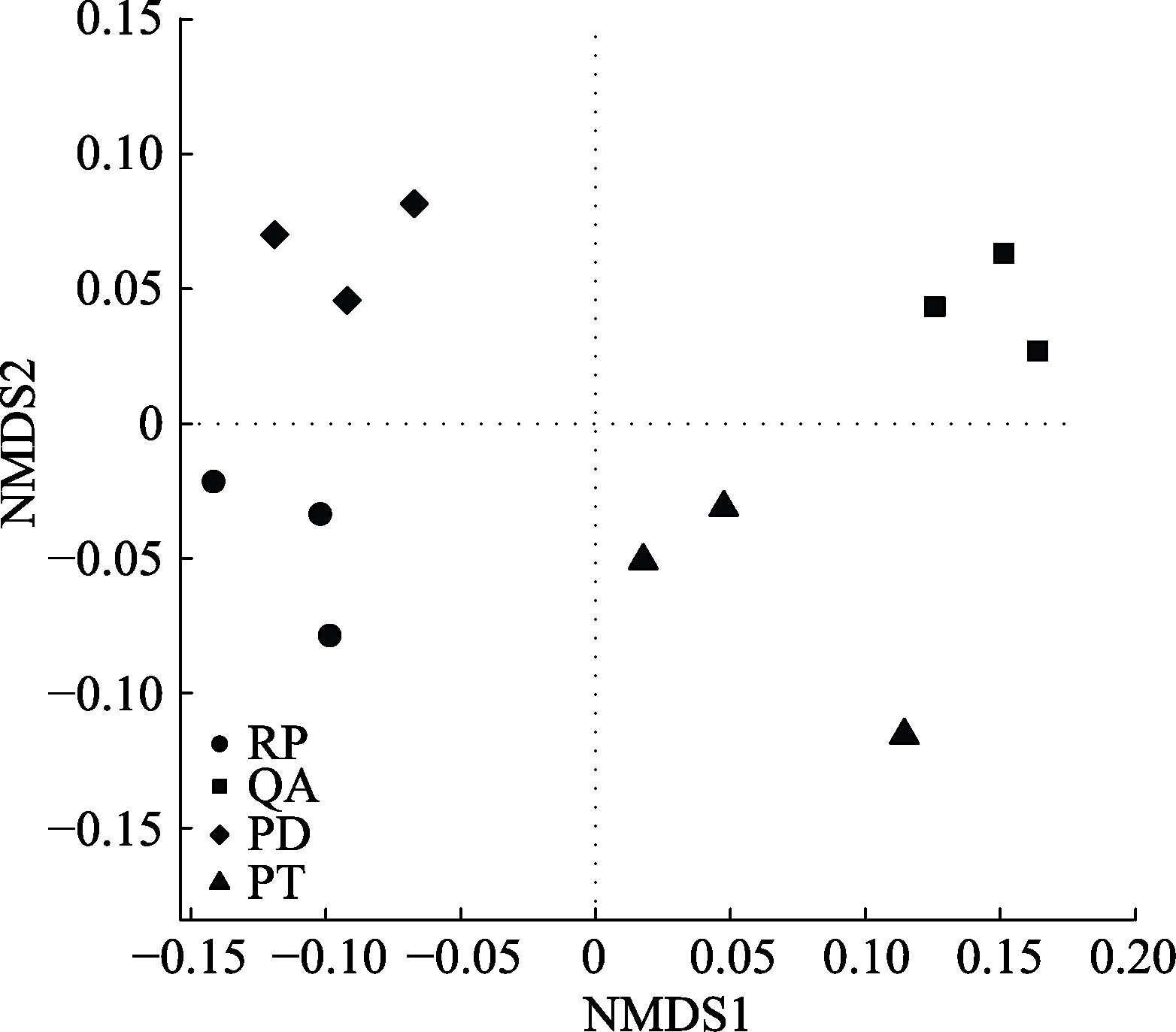
Fig. 2 Nonmetric Multidimensional Scaling (NMDS) ordination diagram of bacterial community structure in root litter after one year of decomposition in Mount Taishan. PD, Pinus densiflora; PT, Pinus tabulaeformis; QA, Quercus acutissima; RP, Robinia pseudoacacia.
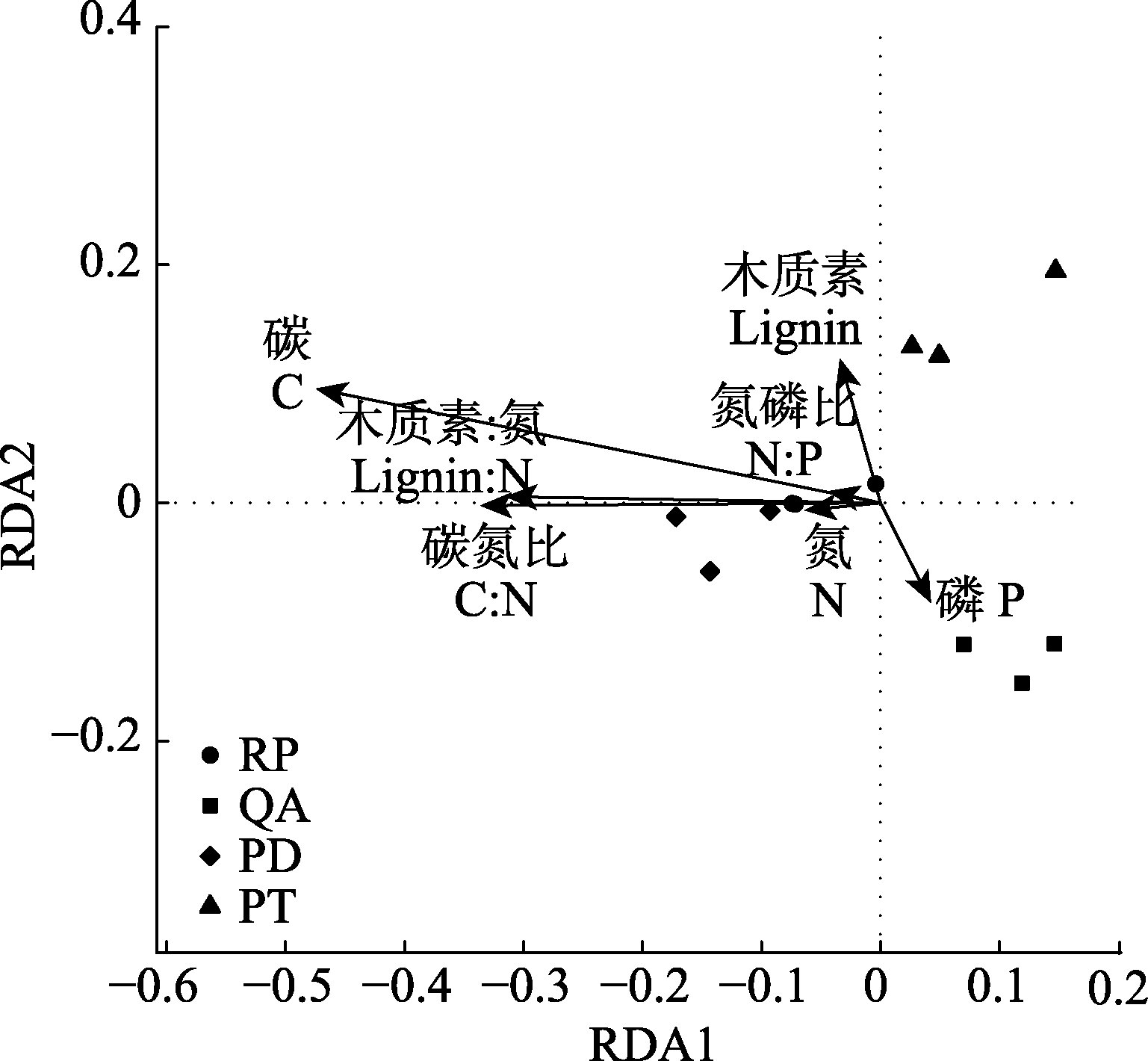
Fig. 3 Redundancy analysis (RDA) based on bacterial community structure and the initial properties of fine root litter. PD, Pinus densiflora; PT, Pinus tabulaeformis; QA, Quercus acutissima; RP, Robinia pseudoacacia.
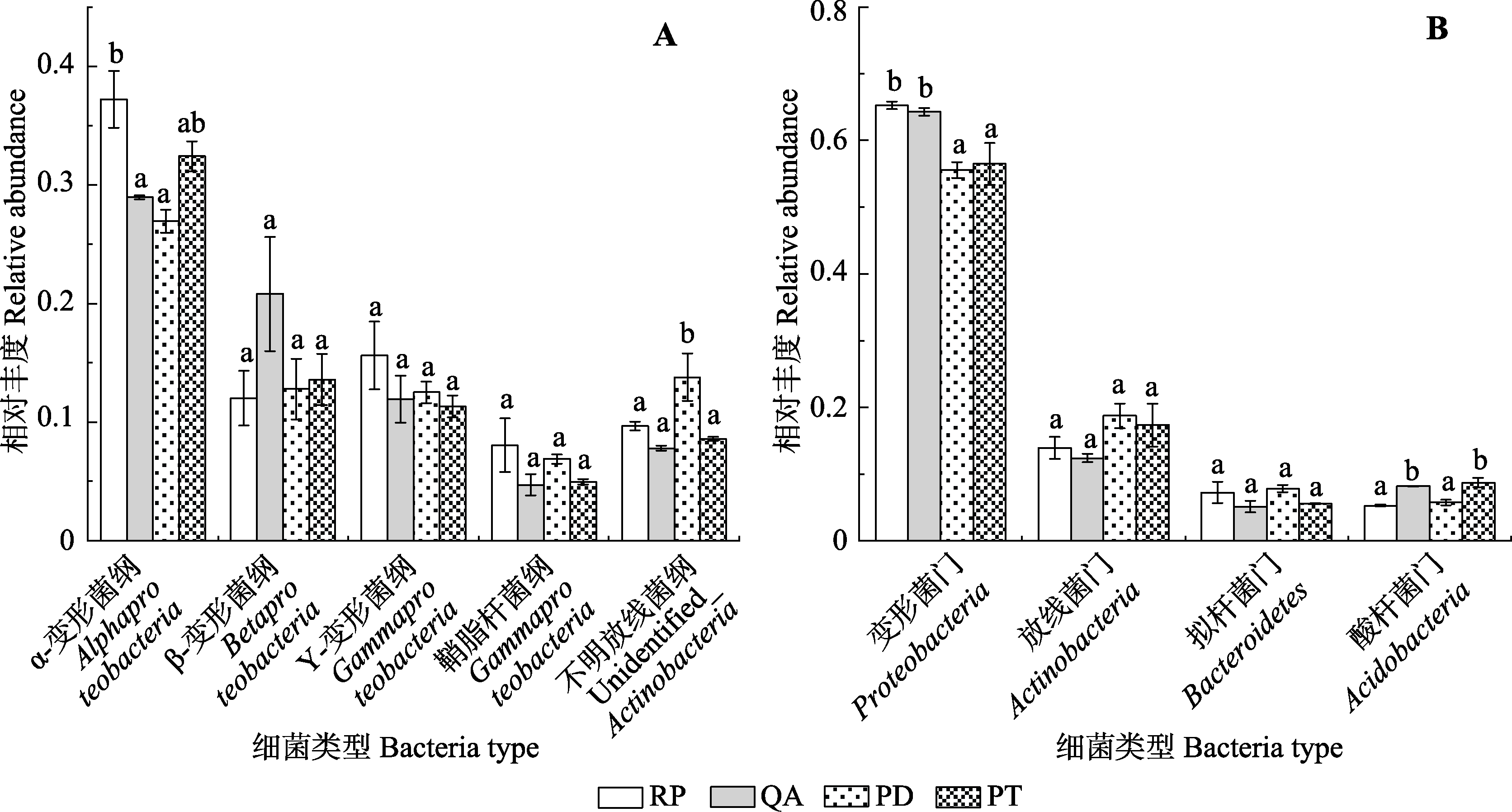
Fig. 4 Differences in relative abundances of major bacterial dominant groups among the four species in Mount Taishan(mean ± SE). A, Dominant classes. B, Dominant phyla. PD, Pinus densiflora; PT, Pinus tabulaeformis; QA, Quercus acutissima; RP, Robinia pseudoacacia. Different lowercase letters indicate the significant differences in different species of the same bacterial group, while the same letter indicates no significant difference.
| 优势门 Dominant phylum | C (%) | N (%) | P (%) | 木质素 Lignin (%) | C:N | N:P | 分解速率 Decomposition rate |
|---|---|---|---|---|---|---|---|
| 变形菌门 Proteobacteria | -0.64 | 0.57 | 0.77* | -0.63 | -0.69* | 0.52 | 0.71* |
| 放线菌门 Actinobacteria | 0.61 | -0.32 | -0.69* | 0.48 | 0.50 | -0.25 | -0.62 |
| 拟杆菌门 Bacteroidetes | 0.60 | 0.09 | 0.09 | 0.09 | 0.43 | 0.03 | -0.36 |
| 酸杆菌门 Acidobacteria | -0.46 | -0.48 | -0.57 | 0.35 | -0.16 | -0.42 | 0.03 |
| 分解速率 Decomposition rate | -0.76** | 0.74** | 0.67* | -0.90** | -0.82** | 0.74** | 1.00 |
Table 4 Correlation analysis among the bacterial dominant phylum , the decomposition rate of fine roots , and the initial properties of litter
| 优势门 Dominant phylum | C (%) | N (%) | P (%) | 木质素 Lignin (%) | C:N | N:P | 分解速率 Decomposition rate |
|---|---|---|---|---|---|---|---|
| 变形菌门 Proteobacteria | -0.64 | 0.57 | 0.77* | -0.63 | -0.69* | 0.52 | 0.71* |
| 放线菌门 Actinobacteria | 0.61 | -0.32 | -0.69* | 0.48 | 0.50 | -0.25 | -0.62 |
| 拟杆菌门 Bacteroidetes | 0.60 | 0.09 | 0.09 | 0.09 | 0.43 | 0.03 | -0.36 |
| 酸杆菌门 Acidobacteria | -0.46 | -0.48 | -0.57 | 0.35 | -0.16 | -0.42 | 0.03 |
| 分解速率 Decomposition rate | -0.76** | 0.74** | 0.67* | -0.90** | -0.82** | 0.74** | 1.00 |
| 优势纲 Dominant class | C (%) | N (%) | P (%) | 木质素 Lignin (%) | C:N | N:P | 分解速率 Decomposition rate |
|---|---|---|---|---|---|---|---|
| α-变形菌纲 Alphaproteobacteria | -0.33 | 0.79** | 0.56 | -0.71* | -0.73* | 0.84** | 0.63* |
| β-变形菌纲 Betaproteobacteria | -0.42 | -0.18 | 0.09 | -0.00 | -0.11 | -0.21 | 0.19 |
| γ-变形菌纲 Gammaproteobacteria | -0.08 | 0.49 | 0.47 | -0.37 | -0.25 | 0.43 | 0.24 |
| 不明放线菌纲unidentified-Actinobacteria | 0.84** | -0.25 | -0.53 | 0.37 | 0.73* | -0.25 | -0.61 |
| 鞘脂杆菌纲 Sphingobacteriia | 0.20 | 0.40 | 0.49 | -0.23 | -0.00 | 0.30 | 0.05 |
Table 5 Correlation analysis among the decomposition rate of fine roots and bacterial dominant class and the initial properties of litter
| 优势纲 Dominant class | C (%) | N (%) | P (%) | 木质素 Lignin (%) | C:N | N:P | 分解速率 Decomposition rate |
|---|---|---|---|---|---|---|---|
| α-变形菌纲 Alphaproteobacteria | -0.33 | 0.79** | 0.56 | -0.71* | -0.73* | 0.84** | 0.63* |
| β-变形菌纲 Betaproteobacteria | -0.42 | -0.18 | 0.09 | -0.00 | -0.11 | -0.21 | 0.19 |
| γ-变形菌纲 Gammaproteobacteria | -0.08 | 0.49 | 0.47 | -0.37 | -0.25 | 0.43 | 0.24 |
| 不明放线菌纲unidentified-Actinobacteria | 0.84** | -0.25 | -0.53 | 0.37 | 0.73* | -0.25 | -0.61 |
| 鞘脂杆菌纲 Sphingobacteriia | 0.20 | 0.40 | 0.49 | -0.23 | -0.00 | 0.30 | 0.05 |
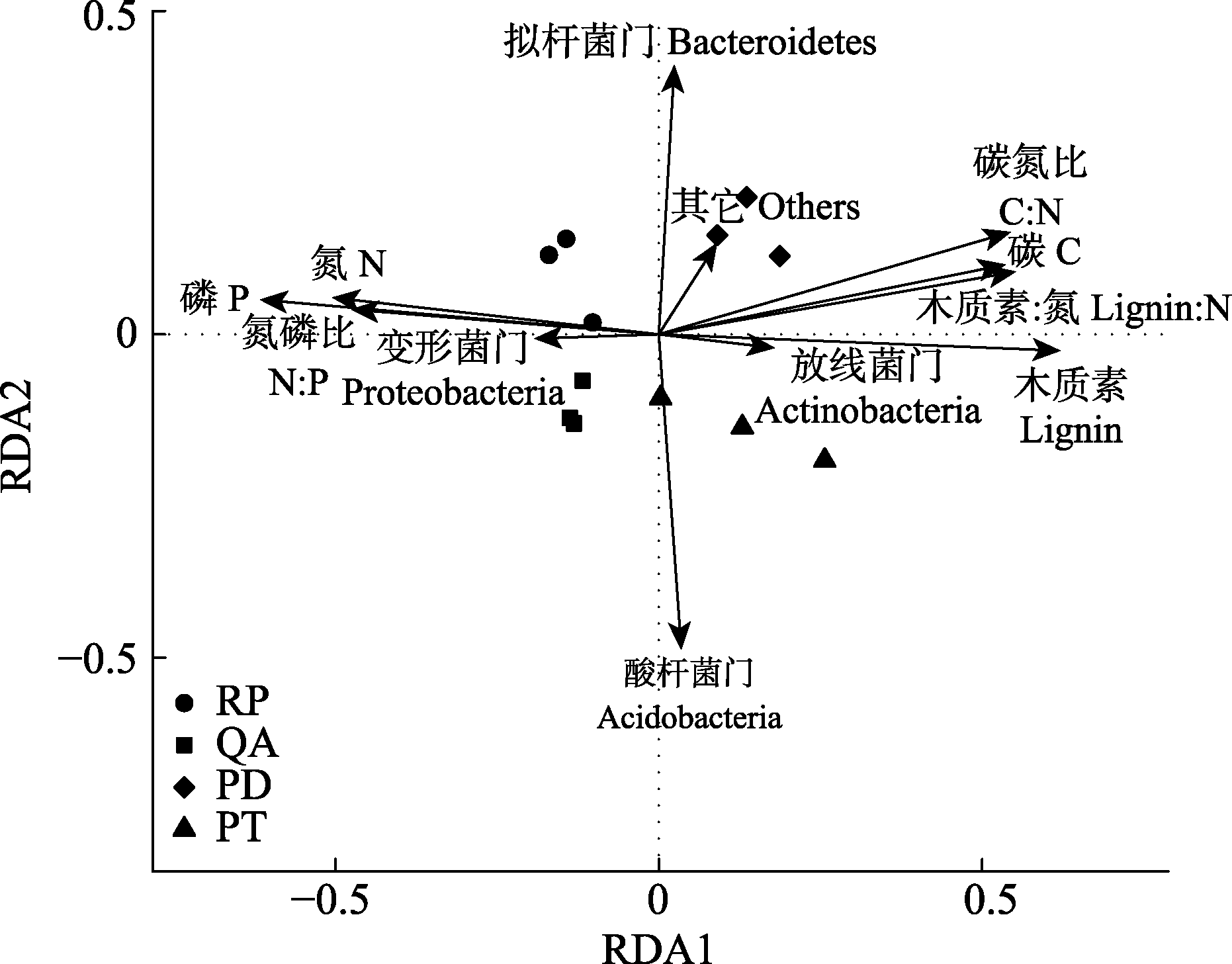
Fig. 5 Redundancy analysis (RDA) based on dominant bacterial phylum and the initial properties of fine root litter. PD, Pinus densiflora; PT, Pinus tabulaeformis; QA, Quercus acutissima; RP, Robinia pseudoacacia.
| [1] |
Adam SW, Zacchaeus GC, Cindy ML ( 2013). Contrasting rRNA gene abundance patterns for aquatic fungi and bacteria in response to leaf-litter chemistry. Freshwater Science, 32, 663-672.
DOI URL |
| [2] |
Barret M, Morrissey JP, O’Gara F ( 2011). Functional genomics analysis of plant growth-promoting rhizobacterial traits involved in rhizosphere competence. Biology and Fertility of Soils, 47, 729-743.
DOI URL |
| [3] |
Chapman SK, Koch GW ( 2007). What type of diversity yields synergy during mixed litter decomposition in a natural forest ecosystem?Plant and Soil, 299, 153-162.
DOI URL |
| [4] |
Chigineva NI, Aleksandrova AV, Tiunov AV ( 2009). The addition of labile carbon alters litter fungal communities and decreases litter decomposition rates. Applied Soil Ecology, 42, 264-270.
DOI URL |
| [5] | Co?teaux M, Bottner P, Berg B ( 1995). Litter decomposition, climate and litter quality. Tree, 10, 63-66. |
| [6] |
Ding XJ, Jing RY, Huang YL, Chen BJ, Ma FY ( 2017). Bacterial structure and diversity in rhizosphere and non-?rhizosphere of Robinia pseudoacacia in the Yellow River Delta. Acta Pedologica Sinica, 54, 1293-1302.
DOI URL |
|
[ 丁新景, 敬如岩, 黄雅丽, 陈博杰, 马风云 ( 2017). 黄河三角洲刺槐根际与非根际细菌结构及多样性. 土壤学报, 54, 1293-1302.]
DOI URL |
|
| [7] |
Eichorst SA, Kuske CR, Schmidt TM ( 2011). Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Applied and Environmental Microbiology, 77, 586-596.
DOI URL PMID |
| [8] |
Elser JJ, Acharya K, Kyle M ( 2003). Growth rate-stoichiometry couplings in diverse biota. Ecology Letters, 6, 936-943.
DOI URL |
| [9] |
Fierer N, Bradford MA, Jackson RB ( 2007). Toward an ecological classification of soil bacteria. Ecology, 88, 1354-1364.
DOI URL |
| [10] |
Gessner MO, Chauvet E, Dobson M ( 1999). A perspective on leaf litter breakdown in streams. Oikos, 85, 377-384.
DOI URL |
| [11] | Grier CC ( 1981). Biomass distribution and above- and below-?ground production in young and mature Abies amabilis zone ecosystems of the Washington Cascades. Canadian Journal of Forest Research, 11, 155-167. |
| [12] |
Gui H, Purahong W, Hyde KD, Xu JC, Mortimer PE ( 2017). The arbuscular mycorrhizal fungus Funneliformis mosseae alters bacterial communities in subtropical forest soils during litter decomposition. Frontiers in Microbiology, 8, 1-11.
DOI URL PMID |
| [13] |
Hultman J, Waldrop MP, Mackelprang R, David MM, McFarland J, Blazewicz SJ, Harden J, Turetsky MR, McGuire AD, Shah MB, VerBerkmoes NC, Lee LH, Mavrommatis K, Jansson JK . ( 2015). Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature, 521, 208-212.
DOI URL PMID |
| [14] |
Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N ( 2009). A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. The ISME Journal, 3, 442-453.
DOI URL PMID |
| [15] |
Kennedy AC ( 1999). Bacterial diversity in agroecosystems. Agriculture Ecosystems and Environment, 74(1-3), 65-76.
DOI URL |
| [16] | Kersters K, De Vos P, Gillis M, Swings J, Vandamme P, Stackebrandt E ( 2006). Introduction to the Proteobacteria. The Prokaryotes, 5(3), 4-37. |
| [17] | Luo YQ, Zhao XY, Wang T, Li YQ, Zuo XA, Ding JP ( 2017). Plant root decomposition and its responses to biotic and abiotic factors. Acta Prataculturae Sinica, 26(2), 197-207. |
| [ 罗永清, 赵学勇, 王涛, 李玉强, 左小安, 丁杰萍 ( 2017). 植物根系分解及其对生物和非生物因素的响应机理研究进展. 草业学报, 26(2), 197-207.] | |
| [18] |
Lydell C, Dowell L, Sikaroodi M, Gillevet P , Emerson ( 2004). A population survey of members of the phylum Bacteroidetes isolated from salt marsh sediments along the east coast of the United States. Microbial Ecology, 48, 263-273.
DOI URL PMID |
| [19] |
McLaren JR, Turkington R ( 2010). Plant functional group identity differentially affects leaf and root decomposition. Global Change Biology, 16, 3075-3084.
DOI URL |
| [20] |
Meentemeyer V ( 1978). Macroclimate and lignin control of litter decomposition rates. Ecology, 59, 465-472.
DOI URL |
| [21] |
Rong L, Li XW, Zhu TH, Zhang J, Yuan WY, Wang Q ( 2009). Varieties of soil microorganisms decomposing Betula luminifera fine roots and Hemarthria compressa roots. Acta Prataculturae Sinica, 18(4), 117-124.
DOI URL |
|
[ 荣丽, 李贤伟, 朱天辉, 张健, 袁渭阳, 王巧 ( 2009). 光皮桦细根与扁穗牛鞭草草根分解的土壤微生物数量及优势类群. 草业学报, 18(4), 117-124.]
DOI URL |
|
| [22] |
Sauvadet M, Chauvat M, Cluzeau D, Maron PA, Villenave C, Bertrand I ( 2016). The dynamics of soil micro-food web structure and functions vary according to litter quality. Soil Biology & Biochemistry, 95, 262-274.
DOI URL |
| [23] | Shen YF, Wang N, Cheng RM, Xiao WF, Yang S, Guo Y, Lei L, Zeng LX, Wang XR ( 2017). Characteristics of fine roots of Pinus massoniana in the Three Gorges Reservoir Area, China. Forests, 8(6), 1-13. |
| [24] |
Soares RA, Roesch LFW, Zanatta G, De Oliveira Camargo FA, Passaglia LMP ( 2006). Occurrence and distribution of nitrogen fixing bacterial community associated with oat ( Avena sativa) assessed by molecular and microbiological techniques. Applied Soil Ecology, 33, 221-234.
DOI URL |
| [25] |
Sun CL, Zhang G, Cheng RJ, Zhu LQ, Ding JB, Chang WS ( 2017). Microbial community structure and diversity of sheepfold atmosphere by 16S rRNA high-throughput sequencing. Acta Veterinaria et Zootechnica Sinica, 48, 1314-1322.
DOI URL |
|
[ 孙翠丽, 张阁, 程汝佳, 朱良全, 丁家波, 常维山 ( 2017). 16S rRNA高通量测序方法检测羊圈空气微生物群落结构及多样性. 畜牧兽医学报, 48, 1314-1322.]
DOI URL |
|
| [26] | Sun H, Wang QX, Liu N, Li L, Zhang CG, Liu ZB, Zhang YY ( 2017). Effects of different leaf litters on the physicochemical properties and bacterial communities in Panax ginseng-growing soil. Applied Soil Ecology, 111, 17-24. |
| [27] |
Taylor BR, Parkinson D, Parsons WFJ ( 1989). Nitrogen and lignin content as predictors of litter decay rates: A microcosm test. Ecology, 70, 97-104.
DOI URL |
| [28] |
Tuomi M, Thum T, Jarvinen H, Fronzek S, Berg B, Harmon M, Trofymow JA, Sevanto S, Liski J ( 2009). Leaf litter decomposition—Estimates of global variability based on Yasso07 model. Ecological Modeling, 220, 3362-3371.
DOI URL |
| [29] |
Urbanová M, ?najdr J, Baldrian P ( 2015). Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biology & Biochemistry, 84, 53-64.
DOI URL |
| [30] |
Větrovsky T, Baldrian P ( 2015). An in-depth analysis of actinobacterial communities shows their high diversity in grassland soils along a gradient of mixed heavy metal contamination. Biology and Fertility of Soils, 51, 827-837.
DOI URL |
| [31] |
Wardle DA, Bardgett RD, Klironomos JN, Set?l? H, van der Putten WH, Wall DH ( 2004). Ecological linkages between aboveground and belowground biota. Science, 304, 1629-1633.
DOI URL PMID |
| [32] |
Wymore AS, Salpas E, Casaburi G, Liu CM, Price LB, Hungate BA, McDowell WH, Marks JC ( 2018). Effects of plant species on stream bacterial communities via leachate from leaf litter. Hydrobiologia, 807, 131-144.
DOI URL |
| [33] |
Zhang CH, Zhang LM, Liu XR, Xin XP, Li SG ( 2011). Root tissue and shoot litter decomposition of dominant species Stipa baicalensis in Hulun Buir meadow steppe of Inner Mongolia, China. Chinese Journal of Plant Ecology, 35, 1156-1166.
DOI URL |
|
[ 张彩虹, 张雷明, 刘杏认, 辛晓平, 李胜功 ( 2011). 呼伦贝尔草甸草原优势种贝加尔针茅根系组织和地上部分凋落物的分解. 植物生态学报, 35, 1156-1166.]
DOI URL |
|
| [34] | Zhang MJ ( 2016). Effects of Forest Gap Disturbance on Microbial Biomass and Bacterial Community Structure in the Process of Foliar Litter Decomposition. Master degree dissertation, Sichuan Agricultural University, Chengdu. |
| [ 张明锦 ( 2016). 林窗干扰对凋落叶分解过程中微生物生物量和细菌群落结构的影响. 硕士论文, 四川农业大学, 成都.] | |
| [35] |
Zhang MJ, Zhang J, Ji TW, Liu H, Li X, Zhang Y, Yang WQ, Chen LH ( 2015). Influence of forest gap on bacterial community structure and diversity during litter decomposition. Ecology and Environment Sciences, 24, 1287-1294.
DOI URL |
|
[ 张明锦, 张健, 纪托未, 刘华, 李勋, 张艳, 杨万勤, 陈良华 ( 2015). 林窗对凋落物分解过程中细菌群落结构和多样性的影响. 生态环境学报, 24, 1287-1294.]
DOI URL |
|
| [36] |
Zhang YG, Cong J, Lu H, Yang CY, Yang YF, Zhou JZ, Li DQ ( 2014). An integrated study to analyze soil microbial community structure and metabolic potential in two forest types. PLOS ONE, 9, e93773. DOI: 10.1371/journal.pone.0093773.
DOI URL PMID |
| [37] |
Zhao BY, Xing P, Wu QL ( 2017). Microbes participated in macrophyte leaf litters decomposition in freshwater habitat. FEMS Microbiology Ecology, 93, fix108. DOI: 10.1093/ femsec/fix108.
DOI URL PMID |
| [38] |
Zhao YY, Wu FZ, Yang WQ, Tan B, He W ( 2016). Variations in bacterial communities during Foliar litter decomposition in the winter and growing seasons in an alpine forest of the eastern Tibetan Plateau. Canadian Journal of Microbiology, 62, 1-14.
DOI URL PMID |
| [1] | Yao Liu Quan-Lin ZHONG Chao-Bin XU Dong-Liang CHENG 芳 跃郑 Zou Yuxing Zhang Xue Xin-Jie Zheng Yun-Ruo Zhou. Relationship between fine root functional traits and rhizosphere microenvironment of Machilus pauhoi at different sizes [J]. Chin J Plant Ecol, 2024, 48(预发表): 0-0. |
| [2] | Ke-Yu CHEN Sen Xing Yu Tang Sun JiaHui Shijie Ren Bao-Ming JI. Arbuscular mycorrhizal fungal community characteristics and driving factors in different grassland types [J]. Chin J Plant Ecol, 2024, 48(5): 660-674. |
| [3] | Xu Zi-Yi Guang-Ze JIN. Variation and trade-offs in fine root functional traits of seedlings of different mycorrhizal types in mixed broadleaved-Korean pine forests [J]. Chin J Plant Ecol, 2024, 48(5): 612-622. |
| [4] | QU Ze-Kun, ZHU Li-Qin, JIANG Qi, WANG Xiao-Hong, YAO Xiao-Dong, CAI Shi-Feng, LUO Su-Zhen, sCHEN Guang-Shui. Nutrient foraging strategies of arbuscular mycorrhizal tree species in a subtropical evergreen broadleaf forest and their relationship with fine root morphology [J]. Chin J Plant Ecol, 2024, 48(4): 416-427. |
| [5] | QIN Wen-Kuan, ZHANG Qiu-Fang, AO Gu-Kai-Lin, ZHU Biao. Responses and mechanisms of soil organic carbon dynamics to warming: a review [J]. Chin J Plant Ecol, 2024, 48(4): 403-415. |
| [6] | YANG An-Na, LI Zeng-Yan, MOU Ling, YANG Bai-Yu, SAI Bi-Le, ZHANG Li, ZHANG Zeng-Ke, WANG Wan-Sheng, DU Yun-Cai, YOU Wen-Hui, YAN En-Rong. Variation in soil bacterial community across vegetation types in Dajinshan Island, Shanghai [J]. Chin J Plant Ecol, 2024, 48(3): 377-389. |
| [7] | NIU Yi-Di, CAI Ti-Jiu. Changes in species diversity and influencing factors in secondary forest succession in northern Da Hinggan Mountains [J]. Chin J Plant Ecol, 2024, 48(3): 349-363. |
| [8] | DU Xu-Long, HUANG Jin-Xue, YANG Zhi-Jie, XIONG De-Cheng. Effects of warming on oxidative damage and defense characteristics and their correlation in leaf and fine root of plants: a review [J]. Chin J Plant Ecol, 2024, 48(2): 135-146. |
| [9] | SHU Wei-Wei, YANG Kun, MA Jun-Xu, MIN Hui-Lin, CHEN Lin, LIU Shi-Ling, HUANG Ri-Yi, MING An-Gang, MING Cai-Dao, TIAN Zu-Wei. Effects of nitrogen addition on the morphological and chemical traits of fine roots with different orders of Castanopsis hystrix [J]. Chin J Plant Ecol, 2024, 48(1): 103-112. |
| [10] | CHEN Zhao-Quan, WANG Ming-Hui, HU Zi-Han, LANG Xue-Dong, HE Yun-Qiong, LIU Wan-De. Mechanisms of seedling community assembly in a monsoon evergreen broadleaf forest in Pu’er, Yunnan, China [J]. Chin J Plant Ecol, 2024, 48(1): 68-79. |
| [11] | LI Na, TANG Shi-Ming, GUO Jian-Ying, TIAN Ru, WANG Shan, HU Bing, LUO Yong-Hong, XU Zhu-Wen. Meta-analysis of effects of grazing on plant community properties in Nei Mongol grassland [J]. Chin J Plant Ecol, 2023, 47(9): 1256-1269. |
| [12] | CHEN Ying-Jie, FANG Kai, QIN Shu-Qi, GUO Yan-Jun, YANG Yuan-He. Spatial patterns and determinants of soil organic carbon component contents and decomposition rate in temperate grasslands of Nei Mongol, China [J]. Chin J Plant Ecol, 2023, 47(9): 1245-1255. |
| [13] | WU Chen, CHEN Xin-Yi, LIU Yuan-Hao, HUANG Jin-Xue, XIONG De-Cheng. Effects of warming on fine root growth, mortality and turnover: a review [J]. Chin J Plant Ecol, 2023, 47(8): 1043-1054. |
| [14] | YANG Xin, REN Ming-Xun. Species distribution pattern and formation mechanism of mangrove plants around the South China Sea [J]. Chin J Plant Ecol, 2023, 47(8): 1105-1115. |
| [15] | YU Xiao, JI Ruo-Xuan, REN Tian-Meng, XIA Xin-Li, YIN Wei-Lun, LIU Chao. Distribution, characteristics and classification of Caryopteris mongholica communities in northern China [J]. Chin J Plant Ecol, 2023, 47(8): 1182-1192. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Chinese Journal of Plant Ecology
Tel: 010-62836134, 62836138, E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn