

植物生态学报 ›› 2025, Vol. 49 ›› Issue (10): 1626-1642.DOI: 10.17521/cjpe.2024.0445 cstr: 32100.14.cjpe.2024.0445
所属专题: 濒危植物种群特征与保护
逯子佳1,2, 王天瑞1, 郑斯斯1, 孟宏虎3,4, 曹建国2, Gregor KOZLOWSKI1,5,6, 宋以刚1,*( )
)
收稿日期:2024-12-09
接受日期:2025-02-07
出版日期:2025-10-20
发布日期:2025-11-20
通讯作者:
*宋以刚(ygsong@cemps.ac.cn)基金资助:
LU Zi-Jia1,2, WANG Tian-Rui1, ZHENG Si-Si1, MENG Hong-Hu3,4, CAO Jian-Guo2, Gregor KOZLOWSKI1,5,6, SONG Yi-Gang1,*( )
)
Received:2024-12-09
Accepted:2025-02-07
Online:2025-10-20
Published:2025-11-20
Supported by:摘要:
气候的快速波动正加速改变着物种命运, 导致部分物种的脆弱性加剧, 并造成许多物种的遗传多样性丧失, 甚至面临灭绝风险。孑遗植物历经新生代以来的极端气候波动, 携带着大量与环境适应相关遗传信息。探讨其种群适应环境的遗传基础及其对未来气候的适应潜力, 可为生物多样性保护提供重要参考依据。该研究以环绕中国四川盆地分布的新生代孑遗植物湖北枫杨(Pterocarya hupehensis)为研究对象, 对其分布范围内18个种群的122个个体进行简化基因组测序; 利用景观基因组学分析方法, 对湖北枫杨的生态适应和遗传脆弱性进行研究。特异性位点检测表明, 398个单核苷酸多态性位点(SNPs)与6个气候因子(等温性、最冷月份最低气温、气温年较差、最湿季度平均气温、最湿月份降水量和降水量季节性变化)具有显著关联, 此外检测到177个受到选择的SNPs位点。梯度森林分析和广义相异模型分析表明降水量季节性变化是影响该物种遗传变异的重要气候因子。Mantel检验检测到显著的环境隔离信号, 冗余分析结果表明环境因素对于遗传变异的解释度大于地理因素。最后, 非适应性风险分析预测到湖北枫杨种群在2090年SSP585情景下的种群脆弱性整体高于SSP126情景, 且降水量季节性变化对于湖北枫杨西北部种群适应能力具有重要影响。该研究不仅为易危物种湖北枫杨在未来气候变化下的管理与保护策略提供了理论依据, 还为环四川盆地孑遗植物如何应对未来气候变化提供新范例。
逯子佳, 王天瑞, 郑斯斯, 孟宏虎, 曹建国, Gregor KOZLOWSKI, 宋以刚. 孑遗植物湖北枫杨的环境适应性遗传变异与遗传脆弱性. 植物生态学报, 2025, 49(10): 1626-1642. DOI: 10.17521/cjpe.2024.0445
LU Zi-Jia, WANG Tian-Rui, ZHENG Si-Si, MENG Hong-Hu, CAO Jian-Guo, Gregor KOZLOWSKI, SONG Yi-Gang. Environmental adaptive genetic variation and genetic vulnerability of relict plant Pterocarya hupehensis. Chinese Journal of Plant Ecology, 2025, 49(10): 1626-1642. DOI: 10.17521/cjpe.2024.0445
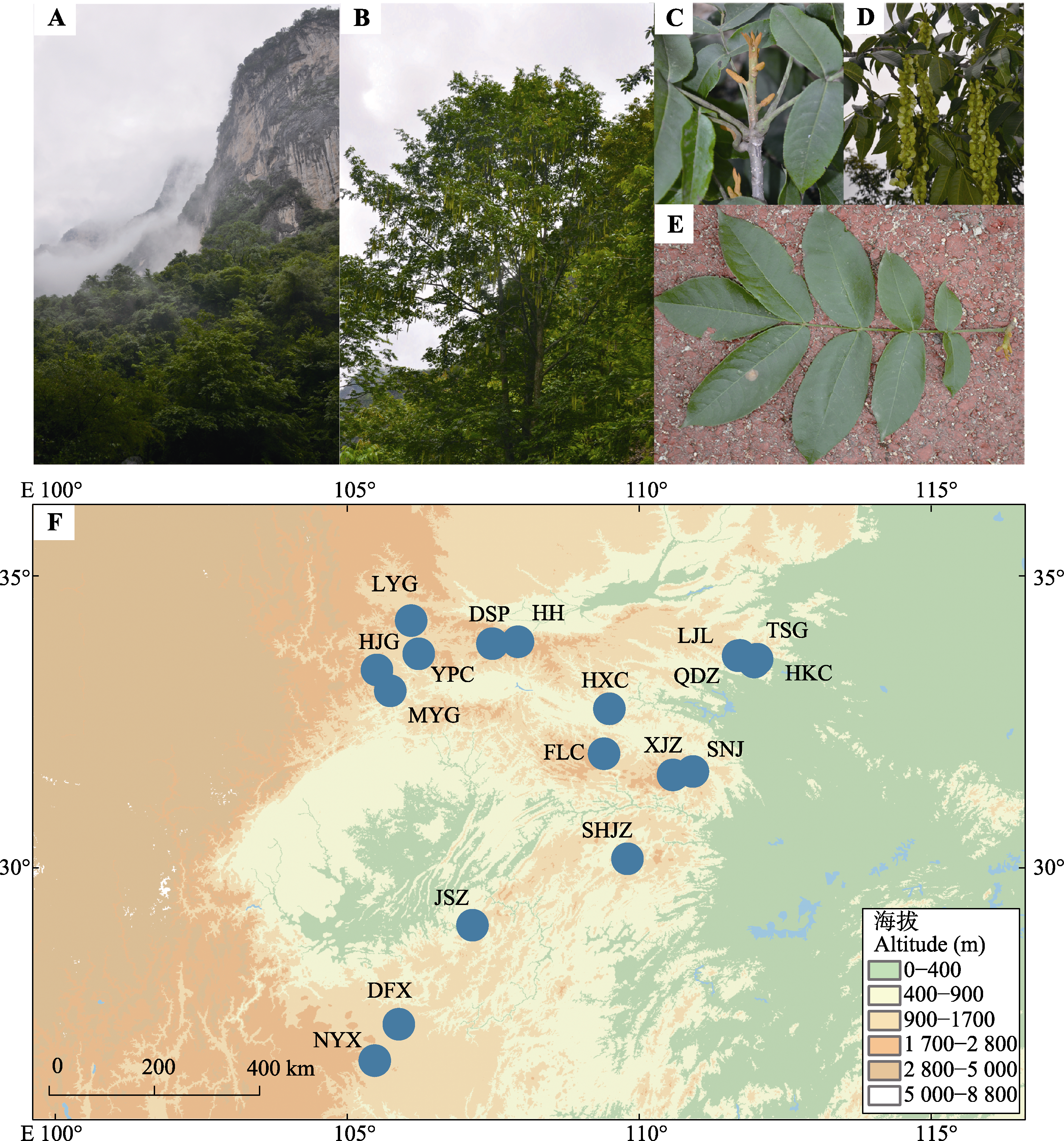
图1 湖北枫杨的生境(A)、植株形态(B-E)和样品采集点(F)。样品采集点信息见表1。
Fig. 1 Habitat of Pterocarya hupehensis (A), plant morphology (B-E), and collection sites of P. hupehensis (F). The information of collection sites is shown in Table 1.
| 种群编号 Code | 采样点 Site | 经度 Longitude (° E) | 纬度 Latitude (° N) | 个体数 n |
|---|---|---|---|---|
| HH | 陕西西安 Xi’an, Shaanxi | 107.93 | 33.87 | 6 |
| HXC | 陕西安康 Ankang, Shaanxi | 109.49 | 32.72 | 3 |
| FLC | 陕西安康 Ankang, Shaanxi | 109.40 | 31.95 | 5 |
| DSP | 陕西宝鸡 Baoji, Shaanxi | 107.49 | 33.84 | 4 |
| QDZ | 河南南阳 Nanyang, Henan | 111.97 | 33.52 | 6 |
| HKC | 河南南阳 Nanyang, Henan | 112.02 | 33.57 | 6 |
| TSG | 河南南阳 Nanyang, Henan | 111.72 | 33.63 | 6 |
| LJL | 河南南阳 Nanyang, Henan | 111.70 | 33.63 | 4 |
| LYG | 甘肃天水 Tianshui, Gansu | 106.10 | 34.23 | 5 |
| YPC | 甘肃陇南 Longnan, Gansu | 106.23 | 33.67 | 5 |
| HJG | 甘肃康县 Kang Xian, Gansu | 105.51 | 33.39 | 5 |
| MYG | 甘肃阳坝镇 Yangba, Gansu | 105.74 | 33.03 | 6 |
| SNJ | 湖北神农架 Shennongjia, Hubei | 110.92 | 31.65 | 7 |
| XJZ | 湖北神农架 Shennongjia, Hubei | 110.58 | 31.59 | 11 |
| SHJZ | 湖北恩施 Enshi, Hubei | 109.80 | 30.16 | 11 |
| JSZ | 重庆金山镇 Jinshan, Chongqing | 107.14 | 29.02 | 7 |
| DFX | 贵州毕节 Bijie, Guizhou | 105.88 | 27.33 | 12 |
| NYX | 贵州毕节 Bijie, Guizhou | 105.47 | 26.70 | 13 |
表1 基于酶切的简化基因组测序数据的18个湖北枫杨种群的样本编码、采样位置以及种群个体数量
Table 1 Sample coding, sampling locations, and individual numbers of 18 Pterocarya hupehensis populations based on restriction-site associated DNA-sequnencing (RAD-seq)
| 种群编号 Code | 采样点 Site | 经度 Longitude (° E) | 纬度 Latitude (° N) | 个体数 n |
|---|---|---|---|---|
| HH | 陕西西安 Xi’an, Shaanxi | 107.93 | 33.87 | 6 |
| HXC | 陕西安康 Ankang, Shaanxi | 109.49 | 32.72 | 3 |
| FLC | 陕西安康 Ankang, Shaanxi | 109.40 | 31.95 | 5 |
| DSP | 陕西宝鸡 Baoji, Shaanxi | 107.49 | 33.84 | 4 |
| QDZ | 河南南阳 Nanyang, Henan | 111.97 | 33.52 | 6 |
| HKC | 河南南阳 Nanyang, Henan | 112.02 | 33.57 | 6 |
| TSG | 河南南阳 Nanyang, Henan | 111.72 | 33.63 | 6 |
| LJL | 河南南阳 Nanyang, Henan | 111.70 | 33.63 | 4 |
| LYG | 甘肃天水 Tianshui, Gansu | 106.10 | 34.23 | 5 |
| YPC | 甘肃陇南 Longnan, Gansu | 106.23 | 33.67 | 5 |
| HJG | 甘肃康县 Kang Xian, Gansu | 105.51 | 33.39 | 5 |
| MYG | 甘肃阳坝镇 Yangba, Gansu | 105.74 | 33.03 | 6 |
| SNJ | 湖北神农架 Shennongjia, Hubei | 110.92 | 31.65 | 7 |
| XJZ | 湖北神农架 Shennongjia, Hubei | 110.58 | 31.59 | 11 |
| SHJZ | 湖北恩施 Enshi, Hubei | 109.80 | 30.16 | 11 |
| JSZ | 重庆金山镇 Jinshan, Chongqing | 107.14 | 29.02 | 7 |
| DFX | 贵州毕节 Bijie, Guizhou | 105.88 | 27.33 | 12 |
| NYX | 贵州毕节 Bijie, Guizhou | 105.47 | 26.70 | 13 |

图2 基于“Pcadapt”的湖北枫杨特异性位点检测结果(A)、主成分分析(PCA)聚类结果(B)和基于潜在因素混合模型分析的对于6个气候因子分别进行特异性位点检测结果(C)。红色虚线为p = 0.01阈值线, 红色虚线上方为与该环境因子显著关联的位点。MAF, 次等位基因频率。
Fig. 2 Abnormal single nucleotide polymorphism (SNP) detected in Pterocarya hupehensis based on “Pcadapt” (A), Principal Component Analysis (PCA) (B), and latent factor mixed models analysis based on six environmental factors (C). The red dotted line represents the threshold of p = 0.01, and the sites above the red dotted line are significantly associated with the environmental factor. MAF, minor allele frequency.
| Mantel检验 Mantel test | Mantel’s r | p | Mantel检验(每个气候因子) Mantel test (Each climatic factor) | Mantel’s r | p |
|---|---|---|---|---|---|
| 地理隔离 Isolation by distance | 0.23 | 0.062 | 等温性 Isothermality | -0.17 | 0.861 |
| 环境隔离 Isolation by environment | 0.31 | 0.002 | 最冷月份最低气温 Minimum temperature of the coldest month | -0.05 | 0.668 |
| 偏Mantel检验 Partial mantel test | 气温年较差 Temperature annual range | -0.07 | 0.675 | ||
| 地理隔离(控制环境距离) Isolation by distance (Conditioned with environmental distance) | 0.08 | 0.298 | 最湿季度平均气温 Mean temperature of the wettest quarter | 0.09 | 0.212 |
| 环境隔离(控制地理距离) Isolation by environment (Conditioned with geographical distance) | 0.23 | 0.029 | 最湿月份降水量 Precipitation of the wettest month | 0.29 | 0.003 |
| 降水量季节性变化 Precipitation seasonality | 0.34 | 0.007 |
表2 湖北枫杨全部种群Mantel和偏Mantel检验结果
Table 2 Mantel and partial Mantel test results for the entire population of Pterocarya hupehensis
| Mantel检验 Mantel test | Mantel’s r | p | Mantel检验(每个气候因子) Mantel test (Each climatic factor) | Mantel’s r | p |
|---|---|---|---|---|---|
| 地理隔离 Isolation by distance | 0.23 | 0.062 | 等温性 Isothermality | -0.17 | 0.861 |
| 环境隔离 Isolation by environment | 0.31 | 0.002 | 最冷月份最低气温 Minimum temperature of the coldest month | -0.05 | 0.668 |
| 偏Mantel检验 Partial mantel test | 气温年较差 Temperature annual range | -0.07 | 0.675 | ||
| 地理隔离(控制环境距离) Isolation by distance (Conditioned with environmental distance) | 0.08 | 0.298 | 最湿季度平均气温 Mean temperature of the wettest quarter | 0.09 | 0.212 |
| 环境隔离(控制地理距离) Isolation by environment (Conditioned with geographical distance) | 0.23 | 0.029 | 最湿月份降水量 Precipitation of the wettest month | 0.29 | 0.003 |
| 降水量季节性变化 Precipitation seasonality | 0.34 | 0.007 |
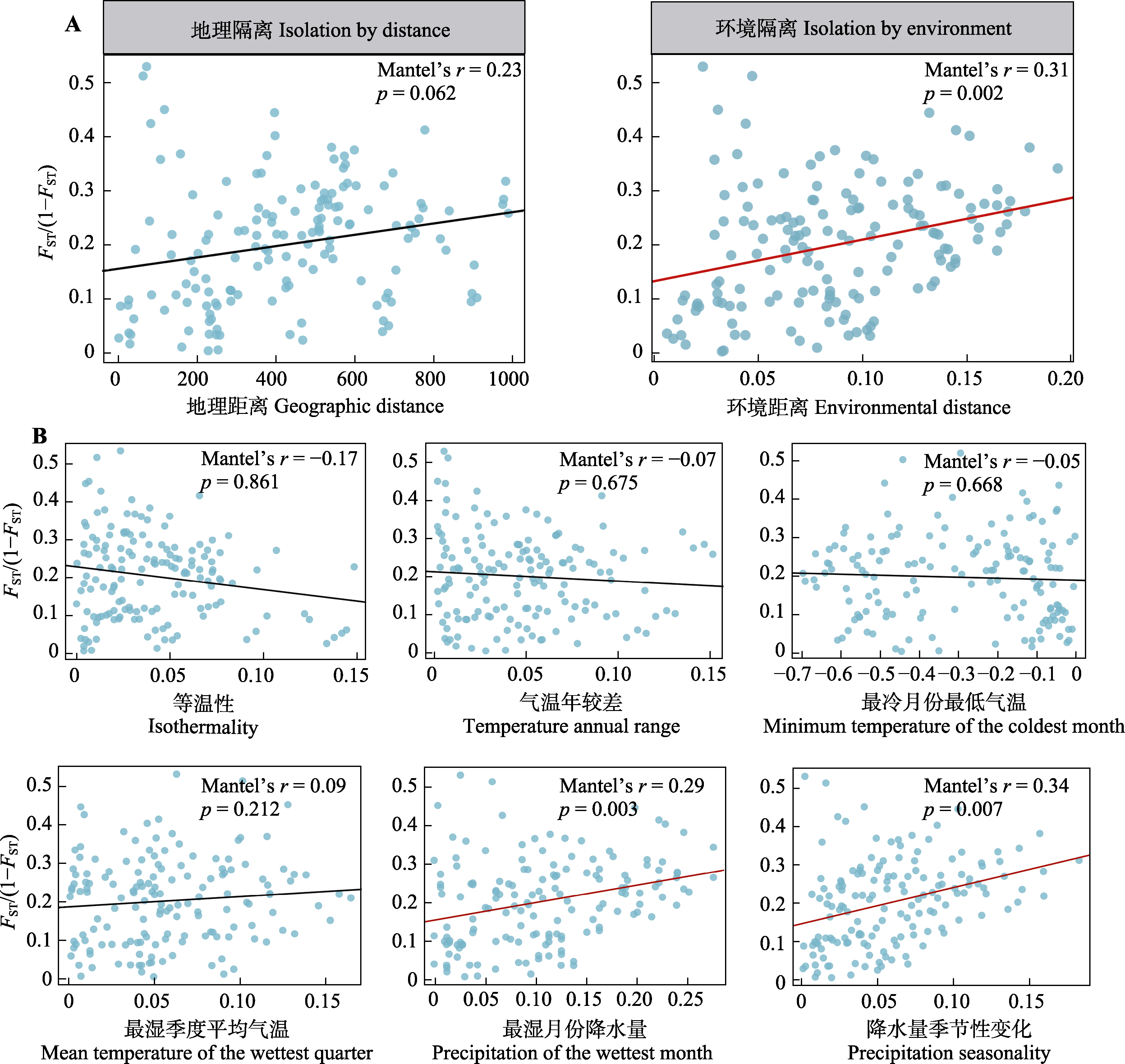
图3 对于所有种群进行地理距离和遗传距离、环境距离和遗传距离的Mantel检验结果(A)和对于每个气候因子进行遗传距离与环境距离检测结果(B)。红色斜线代表变量之间显著相关。FST/(1 - FST), 衡量成对种群间遗传分化(遗传距离)。
Fig. 3 Mantel test results showing the relationship between geographic distance and genetic distance, and between environmental distance and genetic distance across all populations (A), as well as the results of the genetic distance versus environmental distance for each individual climate factor (B). Red dashed lines represent significant correlations between variables. FST/(1 - FST), measuring genetic differentiation between paired populations (genetic distance).

图4 湖北枫杨的遗传变异与6种气候因子(A)及地理因素(B)的冗余分析(RDA)和偏冗余分析(pRDA) (C、D)。向量长度表示该环境变量在解释方差中的贡献大小, 箭头之间的角度表示变量之间的相关性。MEM, 莫兰特征向量。bio3, 等温性; bio6, 最冷月份最低气温; bio7, 气温年较差; bio8, 最湿季度平均气温; bio13, 最湿月份降水量; bio15, 降水量季节性变化。
Fig. 4 Redundancy analysis (RDA) of P. hupehensis between genetic variation and six climatic factors (A), and between genetic variation and geographical factors (B). As well as partial redundancy analysis (pRDA) between genetic variation and six climatic factors (C), and between genetic variation and geographical factors (D). The length of the vector represents the contribution of the environment variable to the explained variance, and the angle between the arrows represents the correlation between the variables. MEM, Moran’s eigenvector. bio3, isothermality; bio6, minimum temperature of the coldest month; bio7, temperature annual range; bio8, mean temperature of the wettest quarter; bio13, precipitation of the wettest month; bio15, precipitation seasonality.
| 环境因子 Environmental factor | 冗余分析 RDA | 偏冗余分析 Partial RDA | ||||
|---|---|---|---|---|---|---|
| PVE | 特征值 Eigenvalue | p | PVE | 特征值 Eigenvalue | p | |
| 地理 Geography | 0.13 | 3.54 | 0.001 | 0.09 | 2.54 | 0.001 |
| 气候 Climate | 0.18 | 4.06 | 0.001 | 0.13 | 3.17 | 0.001 |
| 等温性 Isothermality | 0.03 | 3.63 | 0.001 | 0.03 | 4.05 | 0.001 |
| 最冷月份最低气温 Minimum temperature of the coldest month | 0.03 | 3.70 | 0.001 | 0.02 | 2.60 | 0.001 |
| 气温年较差 Temperature annual range | 0.04 | 5.62 | 0.001 | 0.02 | 2.92 | 0.001 |
| 最湿季度平均气温 Mean temperature of the wettest quarter | 0.02 | 2.82 | 0.001 | 0.03 | 4.24 | 0.001 |
| 最湿月份降水量 Precipitation of the wettest month | 0.05 | 6.71 | 0.001 | 0.01 | 1.81 | 0.001 |
| 降水量季节性变化 Precipitation seasonality | 0.01 | 1.86 | 0.003 | 0.02 | 3.44 | 0.001 |
表3 湖北枫杨所有种群冗余分析和偏冗余分析结果
Table 3 Redundancy analysis (RDA) and partial RDA analysis results of all populations of Pterocarya hupehensis
| 环境因子 Environmental factor | 冗余分析 RDA | 偏冗余分析 Partial RDA | ||||
|---|---|---|---|---|---|---|
| PVE | 特征值 Eigenvalue | p | PVE | 特征值 Eigenvalue | p | |
| 地理 Geography | 0.13 | 3.54 | 0.001 | 0.09 | 2.54 | 0.001 |
| 气候 Climate | 0.18 | 4.06 | 0.001 | 0.13 | 3.17 | 0.001 |
| 等温性 Isothermality | 0.03 | 3.63 | 0.001 | 0.03 | 4.05 | 0.001 |
| 最冷月份最低气温 Minimum temperature of the coldest month | 0.03 | 3.70 | 0.001 | 0.02 | 2.60 | 0.001 |
| 气温年较差 Temperature annual range | 0.04 | 5.62 | 0.001 | 0.02 | 2.92 | 0.001 |
| 最湿季度平均气温 Mean temperature of the wettest quarter | 0.02 | 2.82 | 0.001 | 0.03 | 4.24 | 0.001 |
| 最湿月份降水量 Precipitation of the wettest month | 0.05 | 6.71 | 0.001 | 0.01 | 1.81 | 0.001 |
| 降水量季节性变化 Precipitation seasonality | 0.01 | 1.86 | 0.003 | 0.02 | 3.44 | 0.001 |
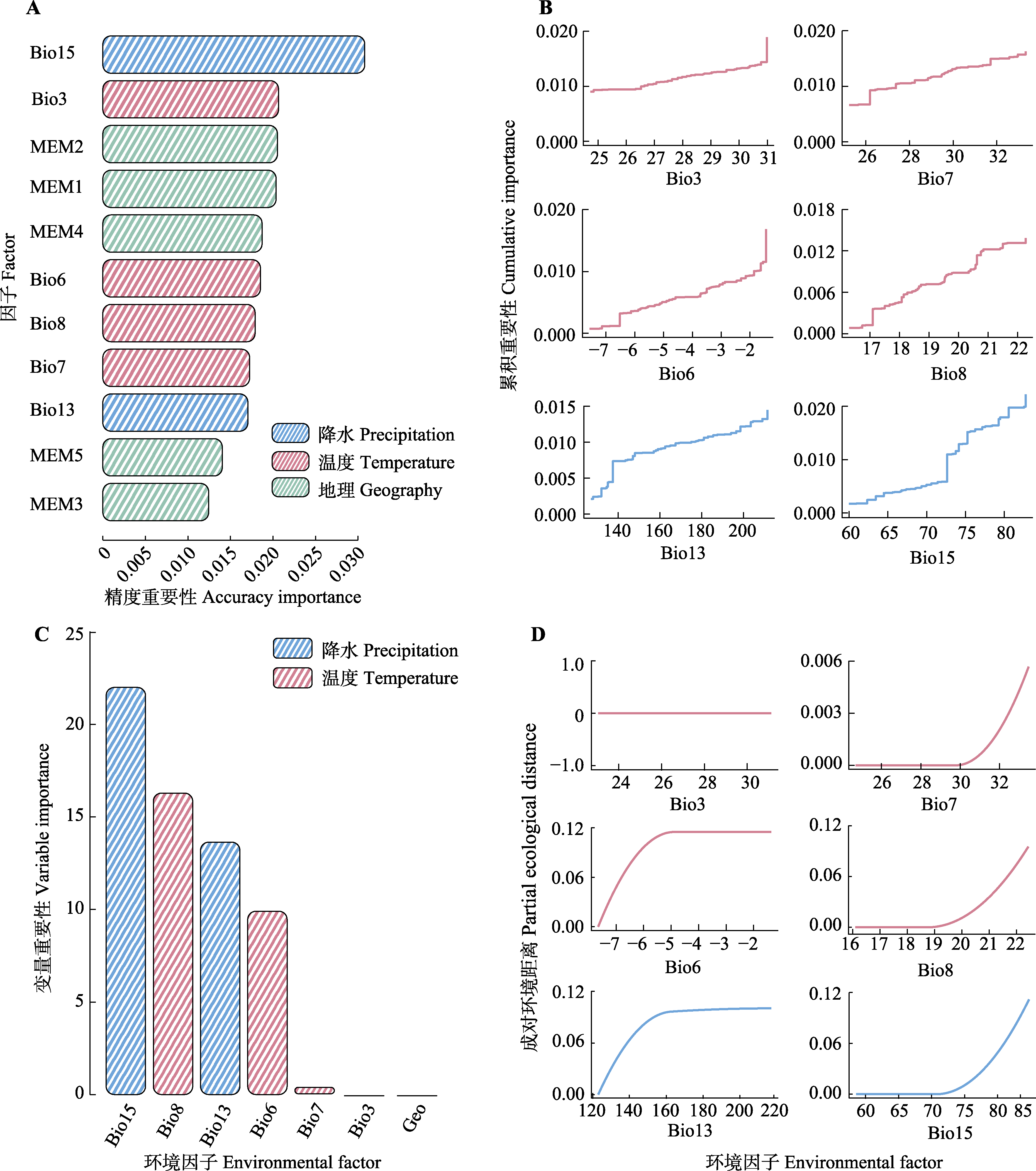
图5 基于梯度森林(GF, A)和广义相异模型(GDM, C)分析的环境变量对于遗传变异重要性排序图, 遗传组成随环境梯度变化的I样条曲线(GF分析结果) (B), 和为GDM分析的累积重要性曲线(D)。MEM, 莫兰特征向量。bio3, 等温性; bio6, 最冷月份最低气温; bio7, 气温年较差; bio8, 最湿季度平均气温; bio13, 最湿月份降水量; bio15, 降水量季节性变化。
Fig. 5 Environmental variable importance rankings for genetic variation based on Gradient Forest (GF) (A) and Generalized Dissimilarity Model (GDM) (C) analyses, with I-spline curves illustrating genetic composition shifts along environmental gradients (B) and cumulative importance curves (D) in GDM analysis. MEM, Moran’s eigenvector. bio3, isothermality; bio6, minimum temperature of the coldest month; bio7, temperature annual range; bio8, mean temperature of the wettest quarter; bio13, precipitation of the wettest month; bio15, precipitation seasonality.
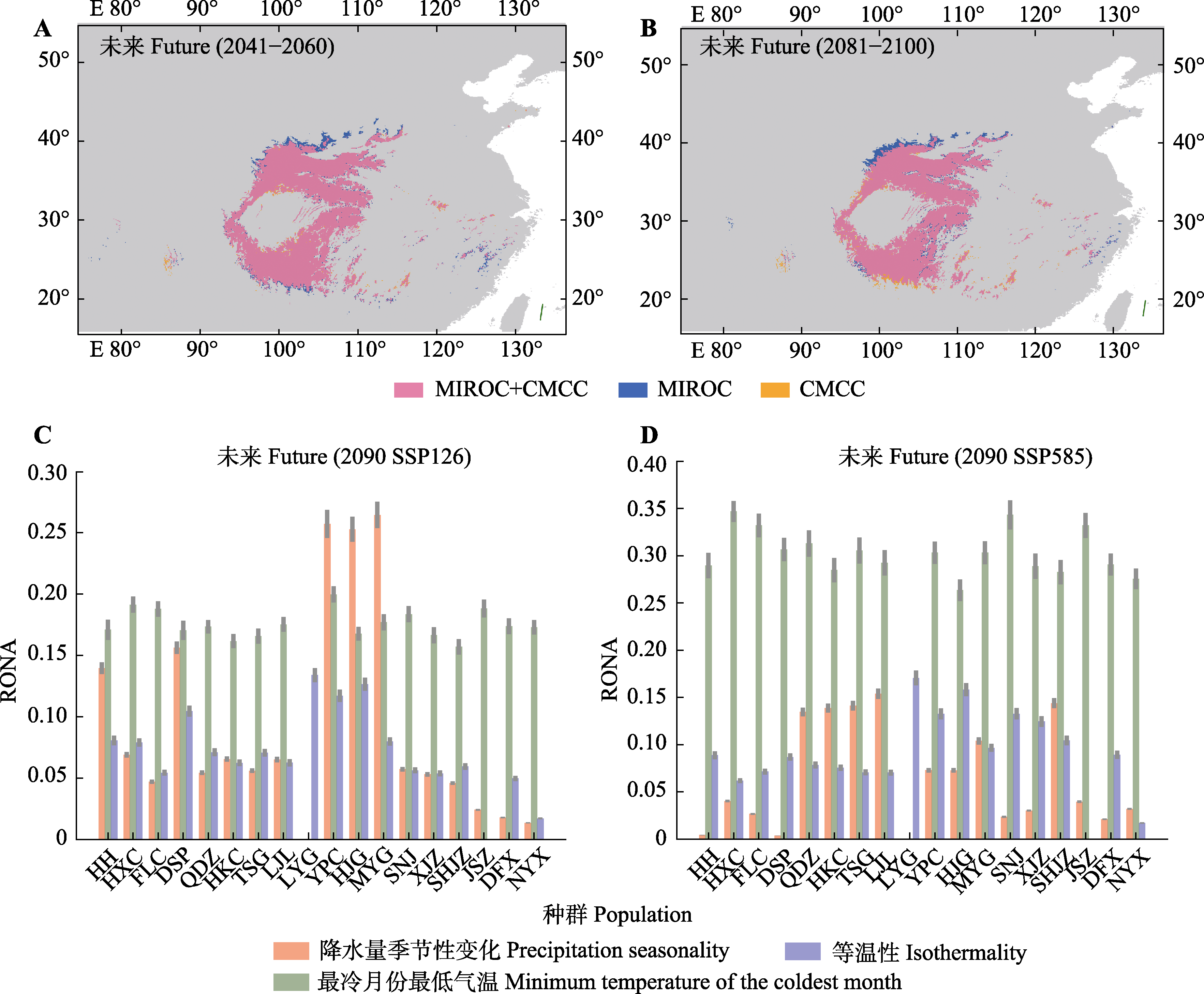
图6 未来气候情景下的潜在分布区预测(A, B)和非适应性风险分析(RONA) (C, D)结果。3个对于基因组脆弱性影响最大的气候因子展示在图中。MIROC和CMCC两种全球气候模型的数据下载于WorldClim v2.1 (https://www.worldclim.org/data/cmip6/ cmip6_clim2.5m.html)。种群名称缩写的详细信息见表1。
Fig. 6 Potential range projections (A, B) and risk of non-adaptedness analysis (RONA) (C, D) for future climate scenarios. Only the three climate factors that have the greatest impact on genomic vulnerability are shown in the picture. Data of two global climate models (MIROC and CMCC) were downloaded from WorldClim v2.1 (https://www.worldclim.org/data/cmip6/cmip6_clim2.5m. html). Detailed information of abbreviations for each population is shown in Table 1.
| [1] |
Aitken SN, Yeaman S, Holliday JA, Wang TL, Curtis-McLane S (2008). Adaptation, migration or extirpation: climate change outcomes for tree populations. Evolutionary Applications, 1, 95-111.
DOI PMID |
| [2] |
Anderson K, Gaston KJ (2013). Lightweight unmanned aerial vehicles will revolutionize spatial ecology. Frontiers in Ecology and the Environment, 11, 138-146.
DOI URL |
| [3] |
Andrews KR, Seaborn T, Egan JP, Fagnan MW, New DD, Chen ZQ, Hohenlohe PA, Waits LP, Caudill CC, Narum SR (2023). Whole genome resequencing identifies local adaptation associated with environmental variation for redband trout. Molecular Ecology, 32, 800-818.
DOI URL |
| [4] |
Bai WN, Wang WT, Zhang DY (2016). Phylogeographic breaks within Asian butternuts indicate the existence of a phytogeographic divide in East Asia. New Phytologist, 209, 1757-1772.
DOI URL |
| [5] | Balkenhol N, Cushman S, Storfer A, Waits L (2015). Landscape Genetics: Concepts, Methods, Applications. Wiley-Blackwell, Oxford, UK. |
| [6] | Balkenhol N, Dudaniec RY, Krutovsky KV, Johnson Jeremy S, Cairns DM, Segelbacher G, Selkoe KA, Heyden VDS, Wang IJ, Selmoni O, Joost Stéphane J (2019). Landscape genomics: understanding relationships between environmental heterogeneity and genomic characteristics of populations//Rajora OP. Population Genomics: Concepts, Approaches and Applications. Springer International Publishing, Cham, Switzerland. 261-322. |
| [7] |
Benito Garzón M, Robson TM, Hampe A (2019). ΔTraitSDMs: species distribution models that account for local adaptation and phenotypic plasticity. New Phytologist, 222, 1757-1765.
DOI PMID |
| [8] |
Cao YN, Comes HP, Sakaguchi S, Chen LY, Qiu YX (2016). Evolution of East Asia’s Arcto-Tertiary relict Euptelea (Eupteleaceae) shaped by Late Neogene vicariance and Quaternary climate change. BMC Evolutionary Biology, 16, 66.
DOI URL |
| [9] |
Capblancq T, Fitzpatrick MC, Bay RA, Exposito-Alonso M, Keller SR (2020). Genomic prediction of (mal)adaptation across current and future climatic landscapes. Annual Review of Ecology, Evolution, and Systematics, 51, 245-269.
DOI |
| [10] |
Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA (2013). Stacks: an analysis tool set for population genomics. Molecular Ecology, 22, 3124-3140.
DOI PMID |
| [11] |
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R,1000 Genomes Project Analysis Group (2011). The variant call format and VCFtools. Bioinformatics, 27, 2156-2158.
DOI PMID |
| [12] |
Dauphin B, Rellstab C, Schmid M, Zoller S, Karger DN, Brodbeck S, Guillaume F, Gugerli F (2021). Genomic vulnerability to rapid climate warming in a tree species with a long generation time. Global Change Biology, 27, 1181-1195.
DOI PMID |
| [13] |
Diniz-Filho JAF, Soares TN, Lima JS, Dobrovolski R, Landeiro VL, de Campos Telles MP, Rangel TF, Bini LM (2013). Mantel test in population genetics. Genetics and Molecular Biology, 36, 475-485.
DOI PMID |
| [14] | Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19, 11-15. |
| [15] |
Ellis N, Smith SJ, Pitcher CR (2012). Gradient forests: calculating importance gradients on physical predictors. Ecology, 93, 156-168.
PMID |
| [16] | Fahad S, Sonmez O, Saud S, Wang D, Wu C, Adnan M, Turan V (2021). Climate Change and Plants: Biodiversity, Growth and Interactions. CRC Press, Boca Raton, USA. |
| [17] |
Ferrier S, Manion G, Elith J, Richardson K (2007). Using generalized dissimilarity modelling to analyse and predict patterns of beta diversity in regional biodiversity assessment. Diversity and Distributions, 13, 252-264.
DOI URL |
| [18] |
Fick SE, Hijmans RJ (2017). WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology, 37, 4302-4315.
DOI URL |
| [19] | Foden WB, Young BE, Akçakaya HR, Garcia RA, Hoffmann AA, Stein BA, Thomas CD, Wheatley CJ, Bickford D, Carr JA, Hole DG, Martin TG, Pacifici M, Pearce-Higgins JW, Platts PJ, et al. (2019). Climate change vulnerability assessment of species. Wiley Interdisciplinary Reviews: Climate Change, 10, e551. DOI: 10.1002/wcc.551. |
| [20] | Frankel OH, Brown AHD, Burdon J (1995). The genetic diversity of wild plants//Volin VC, Volin J. The Conservation of Plant Biodiversity. Cambridge University Press, Cambridge, UK. 10-38. |
| [21] |
Frichot E, François O (2015). LEA: an R package for landscape and ecological association studies. Methods in Ecology and Evolution, 6, 925-929.
DOI URL |
| [22] | Goslee SC, Urban DL (2007). The ecodist package for dissimilarity-based analysis of ecological data. Journal of Statistical Software, 22, 1-19. |
| [23] |
Gougherty AV, Keller SR, Fitzpatrick MC (2021). Maladaptation, migration and extirpation fuel climate change risk in a forest tree species. Nature Climate Change, 11, 166-171.
DOI |
| [24] |
Gugger PF, Liang CT, Sork VL, Hodgskiss P, Wright JW (2018). Applying landscape genomic tools to forest management and restoration of Hawaiian koa (Acacia koa) in a changing environment. Evolutionary Applications, 11, 231-242.
DOI PMID |
| [25] |
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005). Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology, 25, 1965-1978.
DOI URL |
| [26] | Hijmans RJ, Phillips S, Leathwick J, Elith J (2017a). dismo: Species distribution modeling. R package version 1. 1-4. [2024-07-02]. https://CRAN.R-project.org/package=dismo. |
| [27] | Hijmans RJ, van Etten J, Cheng J, Mattiuzzi M, Sumner M, Greenberg JA, Lamigueiro OP, Bevan A, Racine EB, Shortridge A, Ghosh A (2017b). Package ‘raster’: geographic data analysis and modeling, v. 2.6-7. [2024-07-08]. https://cran.r-project.org/web/packages/raster/raster.pdf. |
| [28] | Hijmans RJ, Williams E, Vennes C, Hijmans MR (2021). Package ‘geosphere’. R package version 1.5-14. [2024-06-01]. https://cran.r-project.org/web/packages/geosphere/index.html. |
| [29] |
Hitchings SP, Beebee TJC (1997). Genetic substructuring as a result of barriers to gene flow in urban Rana temporaria (common frog) populations: implications for biodiversity conservation. Heredity, 79, 117-127.
DOI |
| [30] |
Holliday JA, Aitken SN, Cooke JEK, Fady B, González-Martínez SC, Heuertz M, Jaramillo-Correa JP, Lexer C, Staton M, Whetten RW, Plomion C (2017). Advances in ecological genomics in forest trees and applications to genetic resources conservation and breeding. Molecular Ecology, 26, 706-717.
DOI PMID |
| [31] | Hurtt GC, Chini L, Sahajpal R, Frolking S, Bodirsky BL, Calvin K, Doelman JC, Fisk J, Fujimori S, Klein Goldewijk K, Hasegawa T, Havlik P, Heinimann A, Humpenöder F, Jungclaus J, et al. (2020). Harmonization of global land use change and management for the period 850-2100 (LUH2) for CMIP6. Geoscientific Model Development, 13, 5425-5464. |
| [32] | Jiang S, Luo MX, Gao RH, Zhang W, Yang YZ, Li YJ, Liao PC (2019). Isolation-by-environment as a driver of genetic differentiation among populations of the only broad-leaved evergreen shrub Ammopiptanthus mongolicus in Asian temperate deserts. Scientific Reports, 9, 12008. DOI: 10.1038/s41598-019-48472-y. |
| [33] |
Kou YX, Cheng SM, Tian S, Li B, Fan DM, Chen YJ, Soltis DE, Soltis PS, Zhang ZY (2016). The antiquity of Cyclocarya paliurus (Juglandaceae) provides new insights into the evolution of relict plants in subtropical China since the late Early Miocene. Journal of Biogeography, 43, 351-360.
DOI URL |
| [34] | Kozlowski G, Sébastien B, Song YG (2018). Wingnuts (Pterocarya) & Walnut Family. Relict Trees: Linking the Past, Present and Future. Natural History Museum Fribourg, Fribourg, Switzerland. |
| [35] | Kozlowski G, Song Y, Bétrisey S (2019). Pterocarya hupehensis. The IUCN Red List of Threatened Species 2019: e. T66816108A152835141. [2024-09-12]. http://dx. doi.org/10.2305/IUCN.UK.2019-3.RLTS.T66816108A152835141.en. |
| [36] | Kuang KR, Li PQ (1979). Flora of China: Volume 22. Science Press, Beijing. |
| [匡可任, 李沛琼 (1979). 中国植物志: 第二十二卷. 科学出版社, 北京.] | |
| [37] |
Lasky JR, des Marais DL, McKay JK, Richards JH, Juenger TE, Keitt TH (2012). Characterizing genomic variation of Arabidopsis thaliana: the roles of geography and climate. Molecular Ecology, 21, 5512-5529.
DOI URL |
| [38] |
Lefèvre F, Boivin T, Bontemps A, Courbet F, Davi H, Durand-Gillmann M, Fady B, Gauzere J, Gidoin C, Karam MJ, Lalagüe H, Oddou-Muratorio S, Pichot C (2014). Considering evolutionary processes in adaptive forestry. Annals of Forest Science, 71, 723-739.
DOI URL |
| [39] |
Legendre P, Anderson MJ (1999). Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecological Monographs, 69, 1-24.
DOI URL |
| [40] |
Li LF, Cushman SA, He YX, Ma XF, Ge XJ, Li JX, Qian ZH, Li Y (2022). Landscape genomics reveals genetic evidence of local adaptation in a widespread tree, the Chinese wingnut (Pterocarya stenoptera). Journal of Systematics and Evolution, 60, 386-397.
DOI URL |
| [41] | Liaw A, Wiener M (2002). Classification and Regression by randomForest. R News, 2, 18-22. |
| [42] |
Lischer HEL, Excoffier L (2012). PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics, 28, 298-299.
DOI PMID |
| [43] | Lu ZJ, Wang TR, Zheng SS, Meng HH, Cao JG, Song YG, Kozlowski G (2024). Phylogeography of Pterocarya hupehensis reveals the evolutionary patterns of a Cenozoic relict tree around the Sichuan Basin. Forestry Research, 4, e008. DOI: 10.48130/forres-0024-0005. |
| [44] |
Luu K, Bazin E, Blum MGB (2017). Pcadapt: an R package to perform genome scans for selection based on principal component analysis. Molecular Ecology Resources, 17, 67-77.
DOI PMID |
| [45] |
Manel S, Schwartz MK, Luikart G, Taberlet P (2003). Landscape genetics: combining landscape ecology and population genetics. Trends in Ecology & Evolution, 18, 189-197.
DOI URL |
| [46] |
Meinshausen M, Nicholls ZRJ, Lewis J, Gidden MJ, Vogel E, Freund M, Beyerle U, Gessner C, Nauels A, Bauer N, Canadell JG, Daniel JS, John A, Krummel PB, Luderer G, et al. (2020). The shared socio-economic pathway (SSP) greenhouse gas concentrations and their extensions to 2500. Geoscientific Model Development, 13, 3571-3605.
DOI |
| [47] |
Meng HH, Gao XY, Song YG, Cao GL, Li J (2021). Biodiversity arks in the anthropocene. Regional Sustainability, 2, 109-115.
DOI |
| [48] |
Naimi B, Hamm NAS, Groen TA, Skidmore AK, Toxopeus AG (2014). Where is positional uncertainty a problem for species distribution modelling? Ecography, 37, 191-203.
DOI URL |
| [49] |
Pacifici M, Foden WB, Visconti P, Watson JEM, Butchart SHM, Kovacs KM, Scheffers BR, Hole DG, Martin TG, Akçakaya HR, Corlett RT, Huntley B, Bickford D, Carr JA, Hoffmann AA, et al. (2015). Assessing species vulnerability to climate change. Nature Climate Change, 5, 215-224.
DOI |
| [50] |
Pettorelli N, Vik JO, Mysterud A, Gaillard JM, Tucker CJ, Stenseth NC (2005). Using the satellite-derived NDVI to assess ecological responses to environmental change. Trends in Ecology & Evolution, 20, 503-510.
DOI URL |
| [51] |
Phillips SJ, Anderson RP, Schapire RE (2006). Maximum entropy modeling of species geographic distributions. Ecological Modelling, 190, 231-259.
DOI URL |
| [52] |
Pina-Martins F, Baptista J, Pappas G, Paulo OS (2019). New insights into adaptation and population structure of cork oak using genotyping by sequencing. Global Change Biology, 25, 337-350.
DOI PMID |
| [53] |
Privé F, Luu K, Vilhjálmsson BJ, Blum MGB (2020). Performing highly efficient genome scans for local adaptation with R package pcadapt version 4. Molecular Biology and Evolution, 37, 2153-2154.
DOI PMID |
| [54] |
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, Daly MJ, Sham PC (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics, 81, 559-575.
DOI PMID |
| [55] |
Raymond M, Rousset F (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity, 86, 248-249.
DOI URL |
| [56] |
Rellstab C, Gugerli F, Eckert AJ, Hancock AM, Holderegger R (2015). A practical guide to environmental association analysis in landscape genomics. Molecular Ecology, 24, 4348-4370.
DOI PMID |
| [57] |
Rellstab C, Zoller S, Walthert L, Lesur I, Pluess AR, Graf R, Bodénès C, Sperisen C, Kremer A, Gugerli F (2016). Signatures of local adaptation in candidate genes of oaks (Quercus spp.) with respect to present and future climatic conditions. Molecular Ecology, 25, 5907-5924.
DOI URL |
| [58] |
Root TL, Price JT, Hall KR, Schneider SH, Rosenzweig C,Alan Pounds J (2003). Fingerprints of global warming on wild animals and plants. Nature, 421, 57-60.
DOI |
| [59] |
Rousset F (2008). Genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Molecular Ecology Resources, 8, 103-106.
DOI URL |
| [60] | Sang YP, Long ZQ, Dan XM, Feng JJ, Shi TT, Jia CF, Zhang XX, Lai Q, Yang GL, Zhang HY, Xu XT, Liu HH, Jiang YZ, Ingvarsson PK, Liu JQ, Mao KS, Wang J (2022). Genomic insights into local adaptation and future climate-induced vulnerability of a keystone forest tree in East Asia. Nature Communications, 13, 6541. DOI: 10.1038/s41467-022-34206-8. |
| [61] |
Savolainen O, Pyhäjärvi T, Knürr T (2007). Gene flow and local adaptation in trees. Annual Review of Ecology, Evolution, and Systematics, 38, 595-619.
DOI URL |
| [62] | Scheffers BR, De Meester L, Bridge TCL, Hoffmann AA, Pandolfi JM, Corlett RT, Butchart SHM, Pearce-Kelly P, Kovacs KM, Dudgeon D, Pacifici M, Rondinini C, Foden WB, Martin TG, Mora C, et al. (2016). The broad footprint of climate change from genes to biomes to people. Science, 354, aaf7671. DOI: 10.1126/science.aaf7671. |
| [63] | Smith SJ, Ellis N, Pitcher CR (2011). Conditional variable importance in R package extendedForest. [2024-06-06]. https://gradientforest.r-forge.r-project.org/Conditional-importance.pdf. |
| [64] |
Sork VL, Waits L (2010). Contributions of landscape genetics-approaches, insights, and future potential. Molecular Ecology, 19, 3489-3495.
DOI URL |
| [65] |
Storfer A, Murphy MA, Evans JS, Goldberg CS, Robinson S, Spear SF, Dezzani R, Delmelle E, Vierling L, Waits LP (2007). Putting the ‘landscape’ in landscape genetics. Heredity, 98, 128-142.
PMID |
| [66] |
Thuiller W, Albert C, Araújo MB, Berry PM, Cabeza M, Guisan A, Hickler T, Midgley GF, Paterson J, Schurr FM, Sykes MT, Zimmermann NE (2008). Predicting global change impacts on plant species’ distributions: future challenges. Perspectives in Plant Ecology, Evolution and Systematics, 9, 137-152.
DOI URL |
| [67] |
Urban MC (2015). Accelerating extinction risk from climate change. Science, 348, 571-573.
DOI PMID |
| [68] |
van Strien MJ, Holderegger R, Van Heck HJ (2015). Isolation-by-distance in landscapes: considerations for landscape genetics. Heredity, 114, 27-37.
DOI PMID |
| [69] |
Wahid A, Gelani S, Ashraf M, Foolad MR (2007). Heat tolerance in plants: an overview. Environmental and Experimental Botany, 61, 199-223.
DOI URL |
| [70] |
Wang IJ, Bradburd GS (2014). Isolation by environment. Molecular Ecology, 23, 5649-5662.
DOI PMID |
| [71] | Wang TR, Feng L, Du F (2021). New approaches for ecological adaptation study: from population genetics to landscape genomics. Scientia Sinica (Vitae), 51, 167-178. |
| [王天瑞, 冯力, 杜芳 (2021). 生态适应研究新方法: 从种群遗传学到景观基因组学. 中国科学: 生命科学, 51, 167-178. | |
| [72] | Wang TR, Meng HH, Wang N, Zheng SS, Jiang Y, Lin DQ, Song YG, Kozlowski G (2023). Adaptive divergence and genetic vulnerability of relict species under climate change: a case study of Pterocarya macroptera. Annals of Botany, 132, 241-254. |
| [73] | Wang TR, Qiu YX (2024). Application and prospects of landscape genomics in conservation biology. Plant Science Journal, 41, 741-750. |
| [王天瑞, 邱英雄 (2024). 景观基因组学方法在保护生物学研究中的应用与前景. 植物科学学报, 41, 741-750.] | |
| [74] |
Yin QY, Fan Q, Li P, Truong D, Zhao WY, Zhou RC, Chen SF, Liao WB (2021). Neogene and Quaternary climate changes shaped the lineage differentiation and demographic history of Fokienia hodginsii (Cupressaceae s.l.), a Tertiary relict in East Asia. Journal of Systematics and Evolution, 59, 1081-1099.
DOI URL |
| [75] | Zhang WP, Cao L, Lin XR, Ding YM, Liang Y, Zhang DY, Pang EL, Renner SS, Bai WN (2022). Dead-end hybridization in walnut trees revealed by large-scale genomic sequence data. Molecular Biology and Evolution, 39, msab308. DOI: 10.1093/molbev/msab308. |
| [1] | 宋垚彬, 董鸣, 于飞海, 叶学华, 刘建. 克隆植物生态学: 响应与效应[J]. 植物生态学报, 2025, 49(7): 999-1037. |
| [2] | 陈凯, 杨艳, 徐玲, 蒋忠华. 滇西南山地球花报春种子形态与萌发沿海拔梯度的变异[J]. 植物生态学报, 2025, 49(7): 1119-1127. |
| [3] | 王雅轩, 王倩, 林倩缇, 张亦嘉, 郑敏, 顾延生. 神农架大九湖6种草本植物叶解剖结构性状对不同水分生境的响应[J]. , 2025, 49(12): 0-. |
| [4] | 冯珊珊, 黄春晖, 唐梦云, 蒋维昕, 白天道. 细叶云南松针叶形态和显微性状地理变异及其环境解释[J]. 植物生态学报, 2023, 47(8): 1116-1130. |
| [5] | 白天道, 余春兰, 甘泽朝, 赖海荣, 杨隐超, 黄厚宸, 蒋维昕. 细叶云南松种实性状变异与地理气象因子的关联[J]. 植物生态学报, 2020, 44(12): 1224-1235. |
| [6] | 吴毅, 刘文耀, 宋亮, 陈曦, 卢华正, 李苏, 石贤萌. 基于林冠塔吊的附生植物生态学研究进展[J]. 植物生态学报, 2016, 40(5): 508-522. |
| [7] | 周晓旋, 蔡玲玲, 傅梅萍, 洪礼伟, 沈英嘉, 李庆顺. 红树植物胎生现象研究进展[J]. 植物生态学报, 2016, 40(12): 1328-1343. |
| [8] | 郭京衡, 曾凡江, 李尝君, 张波. 塔克拉玛干沙漠南缘三种防护林植物根系构型及其生态适应策略[J]. 植物生态学报, 2014, 38(1): 36-44. |
| [9] | 覃凤飞,李强,崔棹茗,李洪萍,杨智然. 越冬期遮阴条件下3个不同秋眠型紫花苜蓿品种叶片解剖结构与其光生态适应性[J]. 植物生态学报, 2012, 36(4): 333-345. |
| [10] | 李林, 魏识广, 黄忠良, 曹洪麟, 莫德清. 猫儿山两种孑遗植物的更新状况和空间分布格局分析[J]. 植物生态学报, 2012, 36(2): 144-150. |
| [11] | 吉乃提汗·马木提, 谭敦炎, 成小军. 一年生短命植物疏齿千里光果实异形性的生态学意义[J]. 植物生态学报, 2011, 35(6): 663-671. |
| [12] | 周艳松, 王立群. 星毛委陵菜根系构型对草原退化的生态适应[J]. 植物生态学报, 2011, 35(5): 490-499. |
| [13] | 谭敦炎, 张洋, 王爱波. 被子植物地下结实和地上/下两型结实的生态适应意义[J]. 植物生态学报, 2010, 34(1): 72-88. |
| [14] | 刘晓风, 谭敦炎. 24种十字花科短命植物的扩散体特征与扩散对策[J]. 植物生态学报, 2007, 31(6): 1019-1027. |
| [15] | 蔡颖, 关保华, 安树青, 申瑞玲, 蒋金辉, 董蕾. 克隆植物乌菱对底泥磷含量及植株密度的表型可塑性响应[J]. 植物生态学报, 2007, 31(4): 599-606. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
Copyright © 2026 版权所有 《植物生态学报》编辑部
地址: 北京香山南辛村20号, 邮编: 100093
Tel.: 010-62836134, 62836138; Fax: 010-82599431; E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
备案号: 京ICP备16067583号-19
![]()