

Chin J Plant Ecol ›› 2023, Vol. 47 ›› Issue (7): 1020-1031.DOI: 10.17521/cjpe.2022.0335
Special Issue: 微生物生态学
• Research Articles • Previous Articles Next Articles
ZHANG Zhong-Fu1,2, WANG Si-Hai1,2,*( ), YANG Wei1, CHEN Jian1,2
), YANG Wei1, CHEN Jian1,2
Received:2022-08-18
Accepted:2023-03-13
Online:2023-07-20
Published:2023-07-21
Contact:
*WANG Si-Hai(Supported by:ZHANG Zhong-Fu, WANG Si-Hai, YANG Wei, CHEN Jian. Response of rhizosphere microbial community structure and functional characteristics to health status of Malania oleifera[J]. Chin J Plant Ecol, 2023, 47(7): 1020-1031.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.plant-ecology.com/EN/10.17521/cjpe.2022.0335
| 采集地点 Sample location | 海拔 Altitude (m) | 生境 Biotope | 生境特征 Habitat characteristic | 采样数量 Sample size |
|---|---|---|---|---|
| 广南县拖董村 Tuodong village, Guangnan County | 1 280 | 自然分布 Natural distribution | 黄壤, 常绿阔叶林 Yellow soil, evergreen broadleaf forest | 健康和非健康植株各6株 6 healthy and 6 non-healthy individuals |
| 广南县岩腊村 Aila village, Guangnan County | 1 270 | 自然分布 Natural distribution | 石灰岩山地, 蒜头果为主的疏林地 Limestone mountain, Malania oleifera sparse woods | 健康和非健康植株各3株 3 healthy and 3 non-healthy individuals |
| 广南县中就村人工基地 Artificial cultivation in Zhongjiu village, Guangnan County | 1 410 | 人工种植 Artificial cultivation | 黄壤, 人工种植疏杉木林地 Yellow soil, artificial cultivation Cunninghamia lanceolata sparse woods | 健康和非健康植株各3株 3 healthy and 3 non-healthy individuals |
| 富宁县木都村 Mudu village, Funing County | 850 | 自然分布 Natural distribution | 赤红壤, 季风常绿阔叶林 Lateritic red soil, monsoon evergreen broadleaf forest | 健康和非健康植株各3株 3 healthy and 3 non-healthy individuals |
| 富宁县那见村 Najian village, Funing County | 550 | 人工种植 Artificial cultivation | 赤红壤, 次生季风常绿阔叶林 Lateritic red soil, secondary monsoon evergreen broadleaf forest | 健康和非健康植株各3株 3 healthy and 3 non-healthy individuals |
Table 1 Sampling site information of Malania oleifera
| 采集地点 Sample location | 海拔 Altitude (m) | 生境 Biotope | 生境特征 Habitat characteristic | 采样数量 Sample size |
|---|---|---|---|---|
| 广南县拖董村 Tuodong village, Guangnan County | 1 280 | 自然分布 Natural distribution | 黄壤, 常绿阔叶林 Yellow soil, evergreen broadleaf forest | 健康和非健康植株各6株 6 healthy and 6 non-healthy individuals |
| 广南县岩腊村 Aila village, Guangnan County | 1 270 | 自然分布 Natural distribution | 石灰岩山地, 蒜头果为主的疏林地 Limestone mountain, Malania oleifera sparse woods | 健康和非健康植株各3株 3 healthy and 3 non-healthy individuals |
| 广南县中就村人工基地 Artificial cultivation in Zhongjiu village, Guangnan County | 1 410 | 人工种植 Artificial cultivation | 黄壤, 人工种植疏杉木林地 Yellow soil, artificial cultivation Cunninghamia lanceolata sparse woods | 健康和非健康植株各3株 3 healthy and 3 non-healthy individuals |
| 富宁县木都村 Mudu village, Funing County | 850 | 自然分布 Natural distribution | 赤红壤, 季风常绿阔叶林 Lateritic red soil, monsoon evergreen broadleaf forest | 健康和非健康植株各3株 3 healthy and 3 non-healthy individuals |
| 富宁县那见村 Najian village, Funing County | 550 | 人工种植 Artificial cultivation | 赤红壤, 次生季风常绿阔叶林 Lateritic red soil, secondary monsoon evergreen broadleaf forest | 健康和非健康植株各3株 3 healthy and 3 non-healthy individuals |

Fig. 1 Healthy and non-healthy plant morphology of Malania oleifera. A, Healthy individual: spreading crown, lush branches and leaves, less dead branches. B, Non-healthy individual: small crown, sparse branches and leaves, more dead branches.
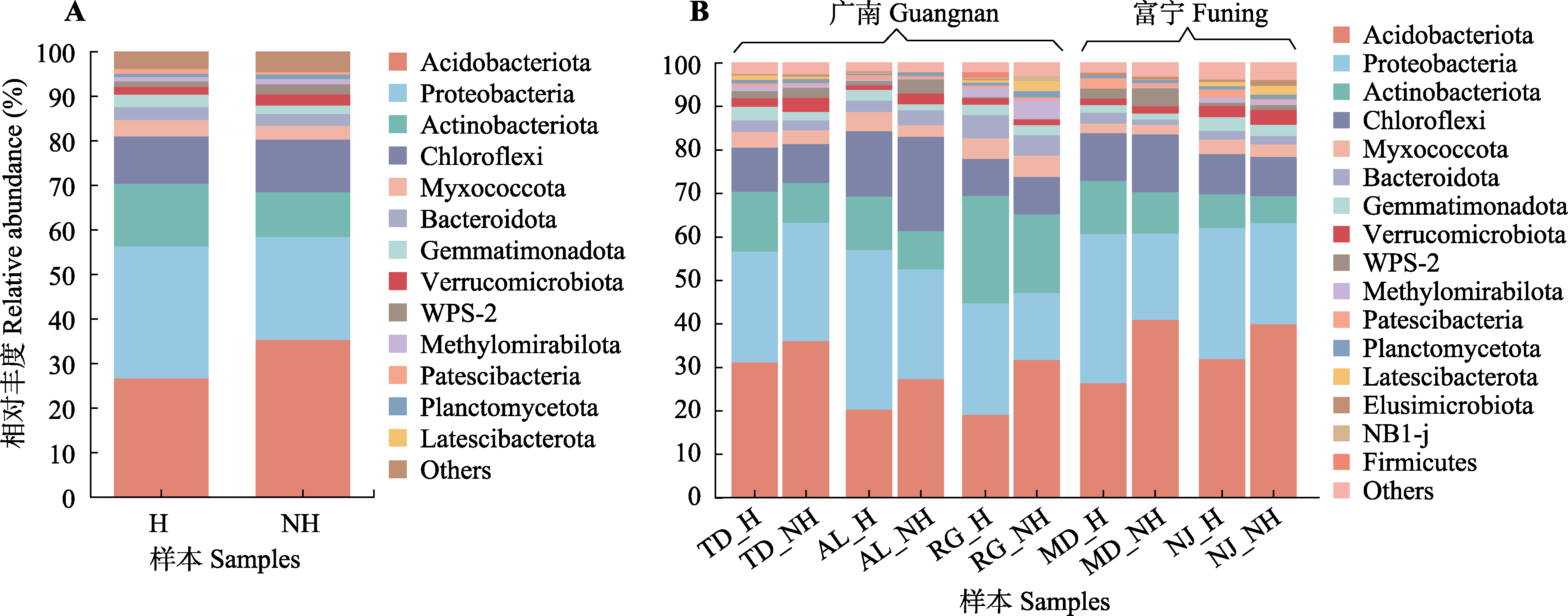
Fig. 2 Root rhizosphere bacteria relative abundance in Malania oleifera. A, Rhizosphere bacteria composition of healthy and non-healthy plants. B, Rhizosphere bacteria composition of five habitats. H, healthy individual; NH, non-healthy individual. AL, Aila village; MD, Mudu village; NJ, Najian village; RG, human planted forest of Zhongjiu village; TD, Tuodong village.
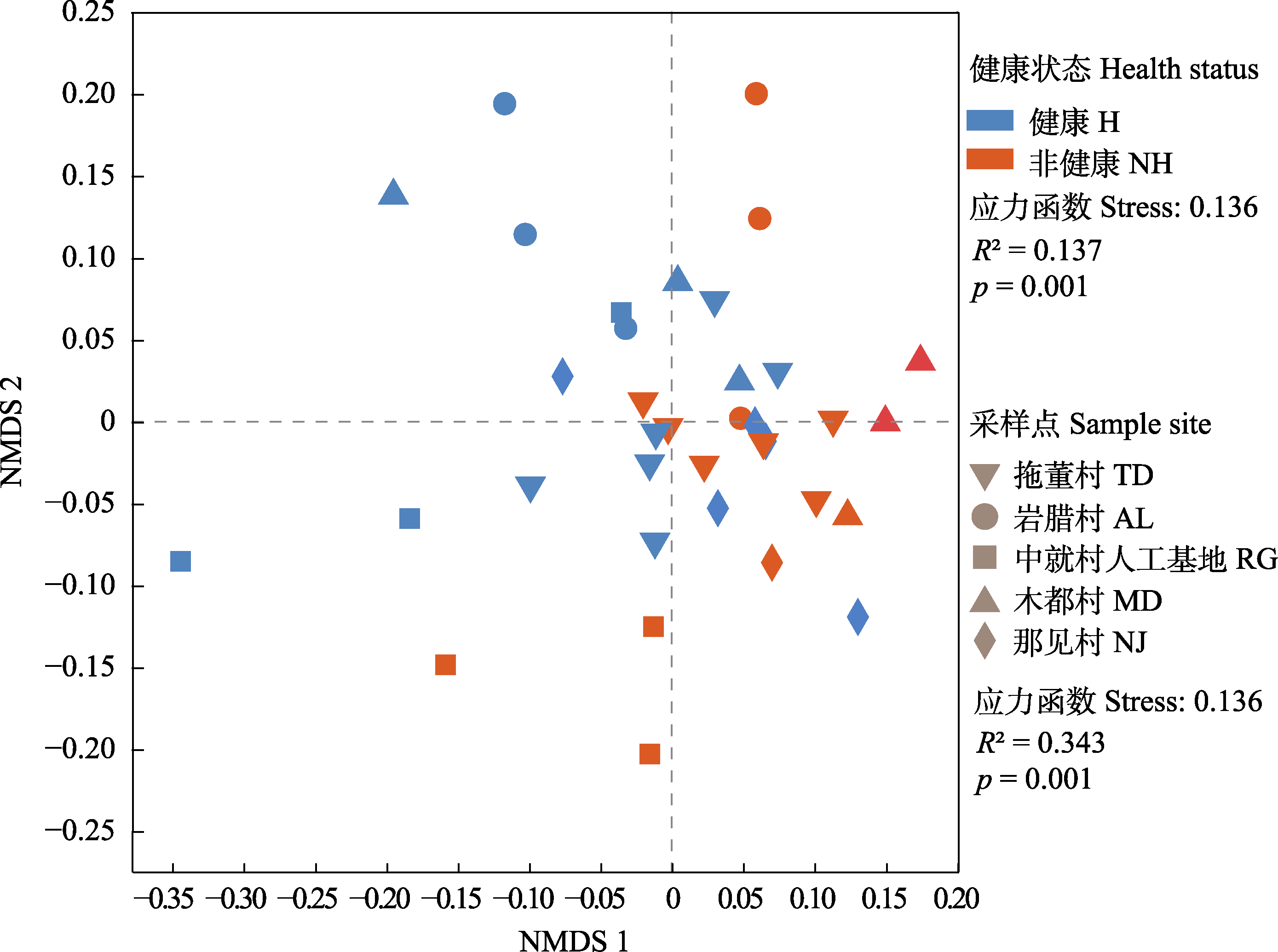
Fig. 3 Root rhizosphere bacteria community of non-metric multidimensional scaling (NMDS) analysis in Malania oleifera. H, healthy individual; NH, non-healthy individual. AL, Aila village; MD, Mudu village; NJ, Najian village; RG, human planted forest of Zhongjiu village; TD, Tuodong village.
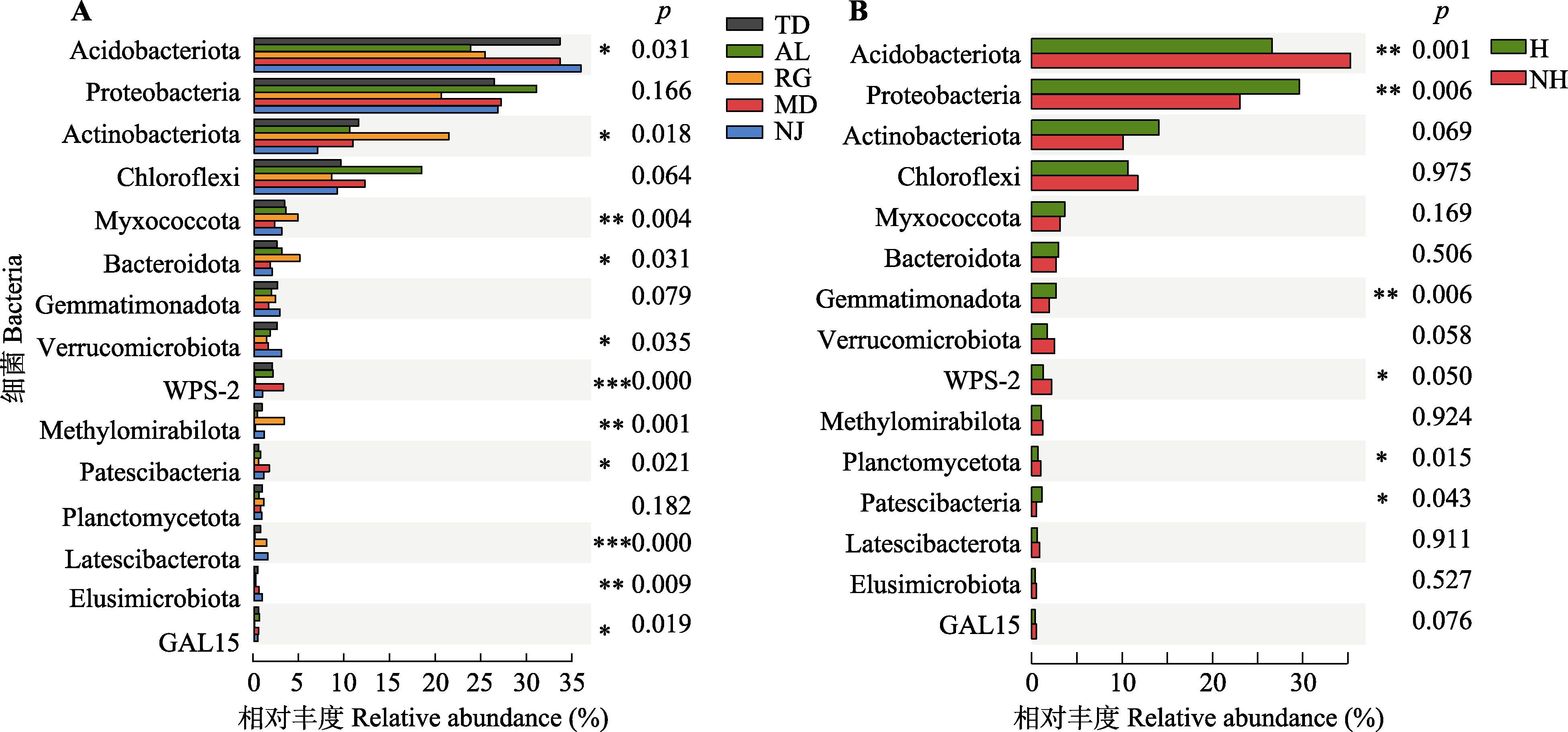
Fig. 4 Difference of root rhizosphere bacteria community in Malania oleifera. A, Bacteria relative abundance in five habitats. B, Healthy and non-healthy individuals in five habitats. *, 0.01 < p ≤ 0.05; **, 0.001 < p ≤ 0.01; ***, p ≤ 0.001. H, healthy individual; NH, non-healthy individual. AL, Aila village; MD, Mudu village; NJ, Najian village; RG, human planted forest of Zhongjiu village; TD, Tuodong village.
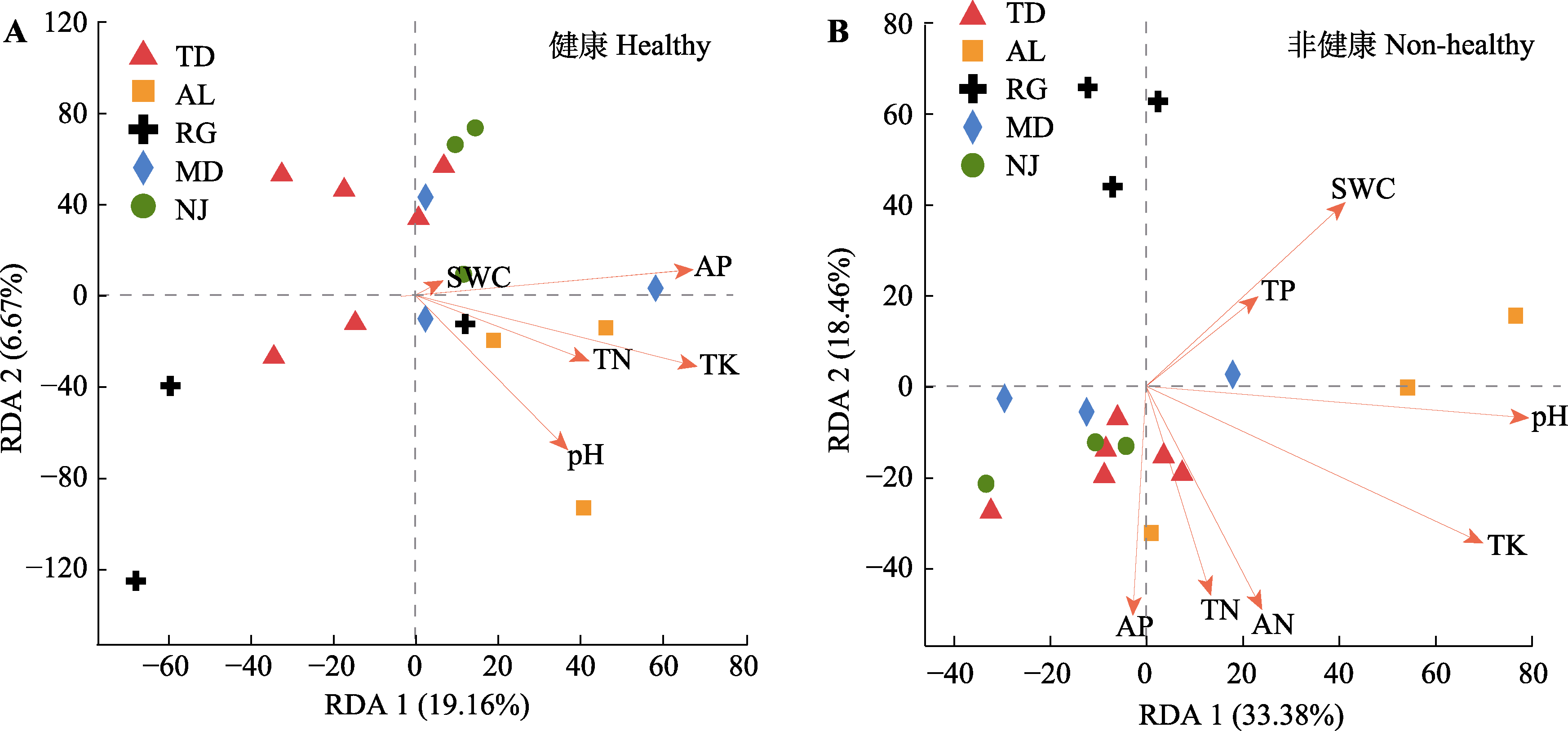
Fig. 5 Rhizosphere bacteria community of redundancy analysis (RDA) in Malania oleifera. A, Healthy individuals in five habitat. B, Non-healthy individuals in five habitats. AN, soil available nitrogen content; AP, soil available phosphorus content; pH, soil pH; SWC, soil water content; TK, soil total potassium content; TN, soil total nitrogen content; TP, soil total phosphorus content. AL, Aila village; MD, Mudu village; NJ, Najian village; RG, human planted forest of Zhongjiu village; TD, Tuodong village.
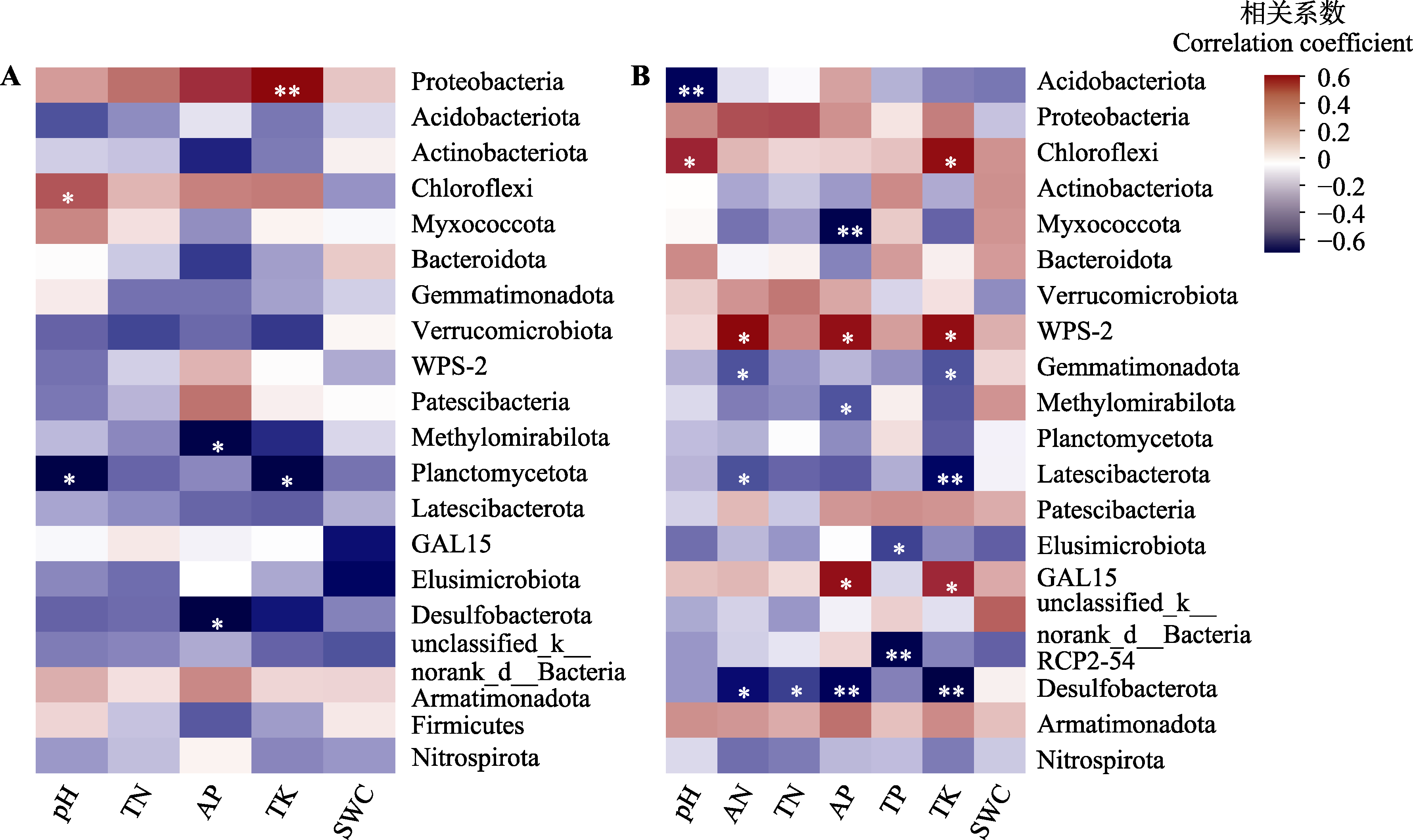
Fig. 6 Pearson correlation heatmap of environmental factors with rhizosphere bacterial taxa in Malania oleifera. A, Healthy individuals. B, Non-healthy individuals. AN, soil available nitrogen content; AP, soil available phosphorus content; pH, soil pH; SWC, soil water content; TK, soil total potassium content; TN, soil total nitrogen content; TP, soil total phosphorus content. *, 0.01 < p ≤ 0.05; **, 0.001 < p ≤ 0.01; ***, p ≤ 0.001.
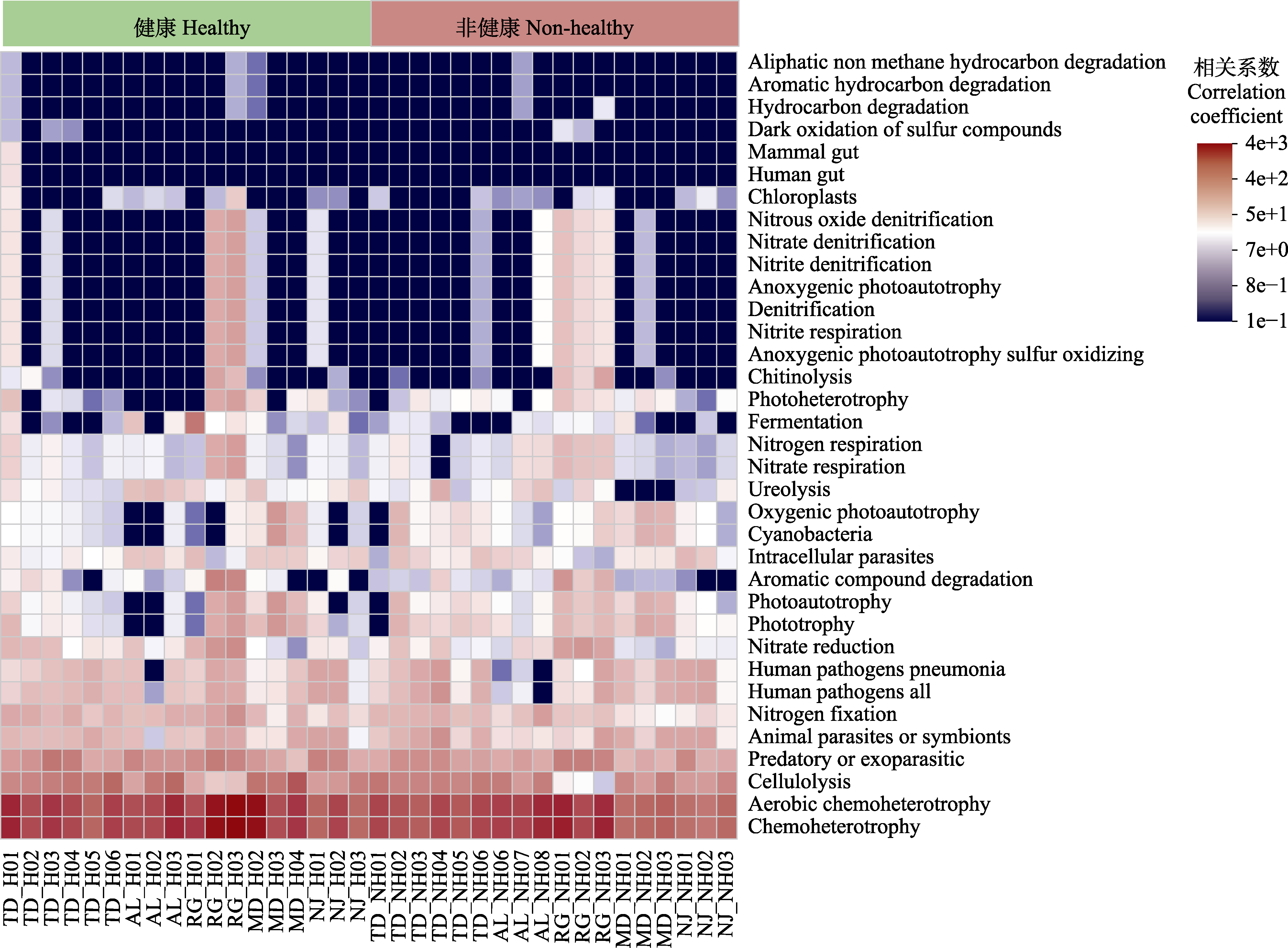
Fig. 7 Pearson correlation heatmap of predicted rhizosphere bacterial function in Malania oleifera. H, healthy individual; NH, non-healthy individual. AL, Aila village; MD, Mudu village; NJ, Najian village; RG, human planted forest of Zhongjiu village; TD, Tuodong village.
| [1] |
Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST, Weigel D, Kemen EM (2016). Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biology, 14, e1002352. DOI: 10.1371/journal.pbio.1002352.
DOI URL |
| [2] |
Andreote FD, de Cássia Pereira e Silva M (2017). Microbial communities associated with plants: learning from nature to apply it in agriculture. Current Opinion in Microbiology, 37, 29-34.
DOI PMID |
| [3] |
Astudillo-García C, Bell JJ, Webster NS, Glasl B, Jompa J, Montoya JM, Taylor MW (2017). Evaluating the core microbiota in complex communities: a systematic investigation. Environmental Microbiology, 19, 1450-1462.
DOI PMID |
| [4] |
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012). The rhizosphere microbiome and plant health. Trends in Plant Science, 17, 478-486.
DOI PMID |
| [5] |
Bouffaud ML, Poirier MA, Muller D, Moënne-Loccoz Y (2014). Root microbiome relates to plant host evolution in maize and other Poaceae. Environmental Microbiology, 16, 2804-2814.
DOI URL |
| [6] |
Chaya A, Kurosawa N, Kawamata A, Kosugi M, Imura S (2019). Community structures of bacteria, archaea, and eukaryotic microbes in the freshwater glacier lake Yukidori-Ike in Langhovde, east Antarctica. Diversity, 11, 105. DOI: 10.3390/d11070105.
DOI |
| [7] |
Chen QL, Ding J, Zhu YG, He JZ, Hu HW (2020a). Soil bacterial taxonomic diversity is critical to maintaining the plant productivity. Environment International, 140, 105766. DOI: 10.1016/j.envint.2020.105766.
DOI URL |
| [8] |
Chen ZM, Wang Q, Ma JW, Chapman S, Zou P, Ye J, Yu QG, Sun WC, Lin H, Jiang LN (2020b). Soil microbial activity and community composition as influenced by application of pig biogas slurry in paddy field in southeast China. Paddy and Water Environment, 18, 15-25.
DOI |
| [9] |
Coleman-Derr D, Desgarennes D, Fonseca-Garcia C, Gross S, Clingenpeel S, Woyke T, North G, Visel A, Partida- Martinez LP, Tringe SG (2016). Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytologist, 209, 798-811.
DOI PMID |
| [10] |
Crossley MN, Dennison WC, Williams RR, Wearing AH (2002). The interaction of water flow and nutrients on aquatic plant growth. Hydrobiologia, 489, 63-70.
DOI URL |
| [11] |
Escudero-Martinez C, Bulgarelli D (2019). Tracing the evolutionary routes of plant-microbiota interactions. Current Opinion in Microbiology, 49, 34-40.
DOI PMID |
| [12] |
Fierer N, Jackson RB (2006). The diversity and biogeography of soil bacterial communities. Proceedings of the National Academy of Sciences of the United States of America, 103, 626-631.
DOI PMID |
| [13] | Fierer N, Leff Jonathan W, Adams Byron J, Nielsen Uffe N, Bates Scott T, Lauber Christian L, Owens S, Gilbert Jack A, Wall Diana H, Caporaso JG (2012). Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proceedings of the National Academy of Sciences of the United States of America, 109, 21390-21395. |
| [14] | Garrido-Oter R, Nakano RT, Dombrowski N, Ma KW, Team A, McHardy AC, Schulze-Lefert P (2018). Modular traits of the rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell Host & Microbe, 24, 155-167. |
| [15] |
Gschwendtner S, Esperschütz J, Buegger F, Reichmann M, Müller M, Munch JC, Schloter M (2011). Effects of genetically modified starch metabolism in potato plants on photosynthate fluxes into the rhizosphere and on microbial degraders of root exudates. FEMS Microbiology Ecology, 76, 564-575.
DOI PMID |
| [16] | Guo FB, Wang SH, Wang J, Zhu F, Chen ZH, Yuan XL (2018). Fruit yield and characters of wild Malania oleifera, a rare plant species in southwest China. Guihaia, 38, 57-64. |
| [郭方斌, 王四海, 王娟, 朱枫, 陈中华, 原晓龙 (2018). 珍稀植物蒜头果野生植株结实量及果实特征研究. 广西植物, 38, 57-64.] | |
| [17] |
Hamonts K, Trivedi P, Garg A, Janitz C, Grinyer J, Holford P, Botha FC, Anderson IC, Singh BK (2018). Field study reveals core plant microbiota and relative importance of their drivers. Environmental Microbiology, 20, 124-140.
DOI PMID |
| [18] |
Hossain Z, Sugiyama SI (2011). Geographical structure of soil microbial communities in northern Japan: effects of distance, land use type and soil properties. European Journal of Soil Biology, 47, 88-94.
DOI URL |
| [19] |
Jacoby R, Peukert M, Succurro A, Koprivova A, Kopriva S (2017). The role of soil microorganisms in plant mineral nutrition—Current knowledge and future directions. Frontiers in Plant Science, 8, 1617. DOI: 10.3389/fpls.2017.01617.
DOI URL |
| [20] | Lai JY (2006). Study on Conservation Biology of Rare and Precious Plant Malania oleifera. PhD dissertation, Sichuang Univerisity, Chengdu. 73-85. |
| [赖家业 (2006). 珍稀植物蒜头果保护生物学研究. 博士学位论文, 四川大学, 成都. 73-85.] | |
| [21] |
Levy A, Salas Gonzalez I, Mittelviefhaus M, Clingenpeel S, Herrera Paredes S, Miao JM, Wang KR, Devescovi G, Stillman K, Monteiro F, Rangel Alvarez B, Lundberg DS, Lu TY, Lebeis S, Jin Z, et al. (2018). Genomic features of bacterial adaptation toplants. Nature Genetics, 50, 138-150.
DOI |
| [22] |
Lewis WH, Tahon G, Geesink P, Sousa DZ, Ettema TJG (2021). Innovations to culturing the uncultured microbial majority. Nature Reviews Microbiology, 19, 225-240.
DOI PMID |
| [23] |
Liu LM, Wang SS, Chen JF (2021a). Anthropogenic activities change the relationship between microbial community taxonomic composition and functional attributes. Environmental Microbiology, 23, 6663-6675.
DOI URL |
| [24] |
Liu ML, Li XR, Zhu RQ, Chen N, Ding L, Chen CY (2021b). Vegetation richness, species identity and soil nutrients drive the shifts in soil bacterial communities during restoration process. Environmental Microbiology Reports, 13, 411-424.
DOI URL |
| [25] | Louca S, Parfrey LW, Doebeli M (2016). Decoupling function and taxonomy in the global ocean microbiome. Science, 35, 1272-1277. |
| [26] |
Lu SP, Gischkat S, Reiche M, Akob DM, Hallberg KB, Küsel K (2010). Ecophysiology of Fe-cycling bacteria in acidic sediments. Applied and Environmental Microbiology, 76, 8174-8183.
DOI PMID |
| [27] |
Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL (2012). Defining the core Arabidopsis thaliana root microbiome. Nature, 488, 86-90.
DOI |
| [28] |
Mendes R, Kruijt M, Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker PAHM, Raaijmakers JM (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science, 332, 1097-1100.
DOI PMID |
| [29] |
Pankratov TA, Kirsanova LA, Kaparullina EN, Kevbrin VV, Dedysh SN (2012). Telmatobacter bradus Gen. nov., sp. nov., a cellulolytic facultative anaerobe from subdivision 1 of the Acidobacteria, and emended description of Acidobacterium capsulatum Kishimoto et al. 1991. International Journal of Systematic and Evolutionary Microbiology, 62, 430-437.
DOI PMID |
| [30] |
Peters NK, Frost JW, Long SR (1986). A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science, 233, 977-980.
DOI PMID |
| [31] |
Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH (2013). Going back to the roots, the microbial ecology of the rhizosphere. Nature Reviews Microbiology, 11, 789-799.
DOI PMID |
| [32] |
Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, van Wees SCM, Bakker PAHM (2014). Induced systemic resistance by beneficial microbes. Annual Review of Phytopathology, 52, 347-375.
DOI PMID |
| [33] |
Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moënne-Loccoz Y (2009). The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant and Soil, 321, 341-361.
DOI URL |
| [34] |
Richardson AE, Barea JM, McNeill AM, Prigent-Combaret C (2009). Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant and Soil, 321, 305-339.
DOI URL |
| [35] |
Rovira AD (1965). Interactions between plant roots and soil microorganisms. Annual Review of Microbiology, 19, 241-266.
PMID |
| [36] |
Saeed Q, Wang XK, Haider FU, Kučerik J, Mumtaz MZ, Holatko J, Naseem M, Kintl A, Ejaz M, Naveed M, Brtnicky M, Mustafa A (2021). Rhizosphere bacteria in plant growth promotion, biocontrol, and bioremediation of contaminated sites: a comprehensive review of effects and mechanisms. International Journal of Molecular Sciences, 22, 10529. DOI: 10.3390/ijms221910529.
DOI URL |
| [37] |
Schulz S, Giebler J, Chatzinotas A, Wick LY, Fetzer I, Welzl G, Harms H, Schloter M (2012). Plant litter and soil type drive abundance, activity and community structure of alkB harbouring microbes in different soil compartments. The ISME Journal, 6, 1763-1774.
DOI |
| [38] |
Trivedi P, Trivedi C, Grinyer J, Anderson IC, Singh BK (2016). Harnessing host-vector microbiome for sustainable plant disease management of phloem-limited bacteria. Frontiers in Plant Science, 7, 1423. DOI: 10.3389/fpls.2016.01423.
DOI PMID |
| [39] |
Vetterlein D, Carminati A, Kögel-Knabner I, Bienert GP, Smalla K, Oburger E, Schnepf A, Banitz T, Tarkka MT, Schlüter S (2020). Rhizosphere spatiotemporal organization—A key to rhizosphere functions. Frontiers in Agronomy, 2, 8. DOI: 10.3389/fagro.2020.00008.
DOI URL |
| [40] |
Vurukonda SSKP, Vardharajula S, Shrivastava M, SkZ A (2016). Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiological Research, 184, 13-24.
DOI PMID |
| [41] |
Wu LK, Lin XM, Lin WX (2014). Advances and perspective in research on plant-soil-microbe interactions mediated by root exudates. Chinese Journal of Plant Ecology, 38, 298-310.
DOI |
|
[吴林坤, 林向民, 林文雄 (2014). 根系分泌物介导下植物-土壤-微生物互作关系研究进展与展望. 植物生态学报, 38, 298-310.]
DOI |
|
| [42] |
Xue PP, Carrillo Y, Pino V, Minasny B, McBratney AB (2018). Soil properties drive microbial community structure in a large scale transect in south eastern Australia. Scientific Reports, 8, 11725. DOI: 10.1038/s41598-018-30005-8.
DOI |
| [43] |
Yang T, Tedersoo L, Soltis PS, Soltis DE, Gilbert JA, Sun M, Shi Y, Wang HF, Li YT, Zhang J, Chen ZD, Lin HY, Zhao YP, Fu CX, Chu HY (2019). Phylogenetic imprint of woody plants on the soil mycobiome in natural mountain forests of eastern China. The ISME Journal, 13, 686-697.
DOI |
| [44] |
York LM, Carminati A, Mooney SJ, Ritz K, Bennett MJ (2016). The holistic rhizosphere, integrating zones, processes, and semantics in the soil influenced by roots. Journal of Experimental Botany, 67, 3629-3643.
DOI URL |
| [45] |
Zhou SB, Peng SC, Li ZQ, Zhang DJ, Zhu YT, Li XQ, Hong MY, Li WC, Lu PL (2022). Characterization of microbial communities and functions in shale gas wastewaters and sludge: implications for pretreatment. Journal of Hazardous Materials, 424, 127649. DOI: 10.1016/j.jhazmat.2021.127649.
DOI URL |
| [1] | Yao Liu Quan-Lin ZHONG Chao-Bin XU Dong-Liang CHENG 芳 跃郑 Zou Yuxing Zhang Xue Xin-Jie Zheng Yun-Ruo Zhou. Relationship between fine root functional traits and rhizosphere microenvironment of Machilus pauhoi at different sizes [J]. Chin J Plant Ecol, 2024, 48(预发表): 0-0. |
| [2] | BAI Xue, LI Yu-Jing, JING Xiu-Qing, ZHAO Xiao-Dong, CHANG Sha-Sha, JING Tao-Yu, LIU Jin-Ru, ZHAO Peng-Yu. Response mechanisms of millet and its rhizosphere soil microbial communities to chromium stress [J]. Chin J Plant Ecol, 2023, 47(3): 418-433. |
| [3] | FENG Ji-Guang, ZHANG Qiu-Fang, YUAN Xia, ZHU Biao. Effects of nitrogen and phosphorus addition on soil organic carbon: review and prospects [J]. Chin J Plant Ecol, 2022, 46(8): 855-870. |
| [4] | NIE Xiu-Qing, WANG Dong, ZHOU Guo-Ying, XIONG Feng, DU Yan-Gong. Soil microbial biomass carbon, nitrogen, phosphorus and their stoichiometric characteristics in alpine wetlands in the Three Rivers Sources Region [J]. Chin J Plant Ecol, 2021, 45(9): 996-1005. |
| [5] | PEI Guang-Ting, SUN Jian-Fei, HE Tong-Xin, HU Bao-Qing. Effects of long-term human disturbances on soil microbial diversity and community structure in a karst grassland ecosystem of northwestern Guangxi, China [J]. Chin J Plant Ecol, 2021, 45(1): 74-84. |
| [6] | LUO Lin, HUANG Yan, LIANG Jin, WANG En-Tao, HU Jun, HE He-Liang, ZHAO Chun-Zhang. Effects of plant interspecific interaction and warming on soil microbial community in root zone soil of two dominant tree species in the subalpine coniferous forest in southwestern China [J]. Chin J Plant Ecol, 2020, 44(8): 875-884. |
| [7] | GAO Gui-Feng, CHU Hai-Yan. Techniques and methods of microbiomics and their applications [J]. Chin J Plant Ecol, 2020, 44(4): 395-408. |
| [8] | WANG Jun, WANG Guan-Qin, LI Fei, PENG Yun-Feng, YANG Gui-Biao, YU Jian-Chun, ZHOU Guo-Ying, YANG Yuan-He. Effects of short-term experimental warming on soil microbes in a typical alpine steppe [J]. Chin J Plan Ecolo, 2018, 42(1): 116-125. |
| [9] | SHI Guo-Xi, WANG Wen-Ying, JIANG Sheng-Jing, CHENG Gang, YAO Bu-Qing, FENG Hu-Yuan, ZHOU Hua-Kun. Effects of the spreading of Ligularia virgaurea on soil physicochemical property and microbial functional diversity [J]. Chin J Plant Ecol, 2018, 42(1): 126-132. |
| [10] | Jiang-Hong ZHANG, Fu-Tian PENG, Xiao-Mei JIANG, Min-Ji LI, Zhong-Tang WANG. Effects of peach branches returning on autotoxins and microbes in soil and tree growth of peaches [J]. Chin J Plan Ecolo, 2016, 40(2): 140-150. |
| [11] | LIANG Ru-Biao,LIANG Jin,QIAO Ming-Feng,XU Zhen-Feng,LIU Qing,YIN Hua-Jun. Effects of simulated exudate C:N stoichiometry on dynamics of carbon and microbial community composition in a subalpine coniferous forest of western Sichuan, China [J]. Chin J Plan Ecolo, 2015, 39(5): 466-476. |
| [12] | WANG Xie,XIANG Cheng-Hua,LI Xian-Wei,WEN Dong-Ju. Short-term effects of a winter wildfire on diversity and intensity of soil microbial function in the subalpine grassland of western Sichuan, China [J]. Chin J Plant Ecol, 2014, 38(5): 468-476. |
| [13] | ZHOU Yong, ZHENG Lu-Yu, ZHU Min-Jie, LI Xia, REN An-Zhi, GAO Yu-Bao. Effects of fungal endophyte infection on soil properties and microbial communities in the host grass habitat [J]. Chin J Plant Ecol, 2014, 38(1): 54-61. |
| [14] | SHI Peng, WANG Shu-Ping, JIA Shu-Gang, GAO Qiang, SUN Xiao-Qiang. Effects of three planting patterns on soil microbial community composition [J]. Chin J Plant Ecol, 2011, 35(9): 965-972. |
| [15] | ZHANG Chong-Bang, WANG Jiang, KE Shi-Xing, JIN Ze-Xin. EFFECTS OF NATURAL INHABITATION BY MISCANTHUS FLORIDULUS ON HEAVY METAL SPECIATIONS AND FUNCTION AND DIVERSITY OF MICROBIAL COMMUNITY IN MINE TAILING SAND [J]. Chin J Plant Ecol, 2009, 33(4): 629-637. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Chinese Journal of Plant Ecology
Tel: 010-62836134, 62836138, E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn