

Chin J Plant Ecol ›› 2022, Vol. 46 ›› Issue (5): 561-568.DOI: 10.17521/cjpe.2021.0354
Special Issue: 青藏高原植物生态学:种群生态学; 生物多样性
• Research Articles • Previous Articles Next Articles
CHEN Tian-Yi1,2, LOU An-Ru1,*( )
)
Received:2021-10-07
Accepted:2022-01-18
Online:2022-05-20
Published:2022-06-09
Contact:
LOU An-Ru
Supported by:CHEN Tian-Yi, LOU An-Ru. Genetic diversity and genetic structure of the Betula platyphylla populations on the eastern side of the Qingzang Plateau[J]. Chin J Plant Ecol, 2022, 46(5): 561-568.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.plant-ecology.com/EN/10.17521/cjpe.2021.0354
| 编号 No. | 采样地点 Sampling location | 种群 Population | 采样数 n | 纬度 Latitude (° N) | 经度 Longitude (° E) | 海拔 Altitude (m) |
|---|---|---|---|---|---|---|
| 1 | 青海省大通县宝库林场 Baoku Forestry Centre, Datong County, Qinghai Province | DA | 33 | 37.14 | 101.56 | 2 714 |
| 2 | 青海省湟源县药水河畔 The riverside of Yaoshui River, Huangyuan County, Qinghai Province | YS | 33 | 36.56 | 101.21 | 2 856 |
| 3 | 甘肃省徽县杂木林中 Miscellaneous wood forest in Hui County, Gansu Province | HX | 29 | 34.08 | 105.73 | 1 518 |
| 4 | 四川省九寨沟县大录乡 Dalu Township, Jiuzhaigou County, Sichuan Province | DL | 31 | 33.57 | 103.67 | 2 600 |
| 5 | 四川省平武县王坝楚 Wangbachu, Pingwu County, Sichuan Province | WB | 35 | 32.74 | 104.32 | 2 130 |
| 6 | 四川省马尔康县城西 West of Barkam County, Sichuan Province | MK | 29 | 31.90 | 102.22 | 2 614 |
| 7 | 四川省红原县刷经寺镇 Shuajingsi Town, Hongyuan County, Sichuan Province | SJ | 32 | 32.02 | 102.61 | 3 309 |
| 8 | 四川省巴塘县G318路旁 G318 roudside, Batang County, Sichuan Province | BT | 34 | 30.30 | 99.39 | 3 521 |
| 9 | 四川省康定县路旁山坡 Roadside slopes, Kangding County, Sichuan Province | KD | 33 | 30.03 | 101.96 | 2 703 |
| 10 | 四川省泸定县杂木林中 Miscellaneous wood forest in Luding County, Sichuan Province | LD | 34 | 29.80 | 102.26 | 2 100 |
| 11 | 云南省德钦县雾浓顶 Wunongding, Dêqên County, Yunnan Province | DN | 29 | 28.45 | 98.91 | 3 585 |
| 12 | 云南省香格里拉市红坡村 Hongpo Village, Xamgyiʹnyilha City, Yunnan Province | HP | 32 | 27.81 | 99.82 | 3 439 |
| 13 | 云南省玉龙县路旁山坡 Roadside slopes, Yulong County, Yunnan Province | YL | 28 | 27.20 | 100.28 | 3 275 |
Table 1 Sampling information of the Betula platyphylla populations on the east side of the Qingzang Plateau
| 编号 No. | 采样地点 Sampling location | 种群 Population | 采样数 n | 纬度 Latitude (° N) | 经度 Longitude (° E) | 海拔 Altitude (m) |
|---|---|---|---|---|---|---|
| 1 | 青海省大通县宝库林场 Baoku Forestry Centre, Datong County, Qinghai Province | DA | 33 | 37.14 | 101.56 | 2 714 |
| 2 | 青海省湟源县药水河畔 The riverside of Yaoshui River, Huangyuan County, Qinghai Province | YS | 33 | 36.56 | 101.21 | 2 856 |
| 3 | 甘肃省徽县杂木林中 Miscellaneous wood forest in Hui County, Gansu Province | HX | 29 | 34.08 | 105.73 | 1 518 |
| 4 | 四川省九寨沟县大录乡 Dalu Township, Jiuzhaigou County, Sichuan Province | DL | 31 | 33.57 | 103.67 | 2 600 |
| 5 | 四川省平武县王坝楚 Wangbachu, Pingwu County, Sichuan Province | WB | 35 | 32.74 | 104.32 | 2 130 |
| 6 | 四川省马尔康县城西 West of Barkam County, Sichuan Province | MK | 29 | 31.90 | 102.22 | 2 614 |
| 7 | 四川省红原县刷经寺镇 Shuajingsi Town, Hongyuan County, Sichuan Province | SJ | 32 | 32.02 | 102.61 | 3 309 |
| 8 | 四川省巴塘县G318路旁 G318 roudside, Batang County, Sichuan Province | BT | 34 | 30.30 | 99.39 | 3 521 |
| 9 | 四川省康定县路旁山坡 Roadside slopes, Kangding County, Sichuan Province | KD | 33 | 30.03 | 101.96 | 2 703 |
| 10 | 四川省泸定县杂木林中 Miscellaneous wood forest in Luding County, Sichuan Province | LD | 34 | 29.80 | 102.26 | 2 100 |
| 11 | 云南省德钦县雾浓顶 Wunongding, Dêqên County, Yunnan Province | DN | 29 | 28.45 | 98.91 | 3 585 |
| 12 | 云南省香格里拉市红坡村 Hongpo Village, Xamgyiʹnyilha City, Yunnan Province | HP | 32 | 27.81 | 99.82 | 3 439 |
| 13 | 云南省玉龙县路旁山坡 Roadside slopes, Yulong County, Yunnan Province | YL | 28 | 27.20 | 100.28 | 3 275 |
| 种群 Population | 等位基因数 AO | 观察杂合度 HO | 期望杂合度 HE | 等位基因丰富度 Ar | 特有等位基因丰富度 PA | 近交系数 Fis |
|---|---|---|---|---|---|---|
| DA | 74 | 0.691 | 0.703 | 6.26 | 0.43 | 0.032 |
| YS | 67 | 0.636 | 0.699 | 5.76 | 0.15 | 0.105* |
| HX | 67 | 0.674 | 0.687 | 5.90 | 0.19 | 0.036 |
| DL | 72 | 0.657 | 0.693 | 6.12 | 0.32 | 0.068* |
| WB | 59 | 0.548 | 0.613 | 5.06 | 0.00 | 0.120* |
| MK | 65 | 0.621 | 0.662 | 5.67 | 0.10 | 0.080* |
| SJ | 65 | 0.676 | 0.674 | 5.59 | 0.08 | 0.013 |
| BT | 49 | 0.455 | 0.478 | 4.12 | 0.07 | 0.063 |
| KD | 62 | 0.612 | 0.588 | 5.23 | 0.42 | -0.025 |
| LD | 47 | 0.591 | 0.575 | 4.13 | 0.01 | -0.013 |
| DN | 33 | 0.334 | 0.324 | 2.91 | 0.00 | -0.012 |
| HP | 43 | 0.364 | 0.403 | 3.67 | 0.21 | 0.114* |
| YL | 36 | 0.361 | 0.430 | 3.20 | 0.00 | 0.178* |
| 平均值 Mean | 57 | 0.555 | 0.579 | 4.89 | 0.15 | - |
Table 2 Genetic diversity of the Betula platyphylla populations on the east side of the Qingzang Plateau
| 种群 Population | 等位基因数 AO | 观察杂合度 HO | 期望杂合度 HE | 等位基因丰富度 Ar | 特有等位基因丰富度 PA | 近交系数 Fis |
|---|---|---|---|---|---|---|
| DA | 74 | 0.691 | 0.703 | 6.26 | 0.43 | 0.032 |
| YS | 67 | 0.636 | 0.699 | 5.76 | 0.15 | 0.105* |
| HX | 67 | 0.674 | 0.687 | 5.90 | 0.19 | 0.036 |
| DL | 72 | 0.657 | 0.693 | 6.12 | 0.32 | 0.068* |
| WB | 59 | 0.548 | 0.613 | 5.06 | 0.00 | 0.120* |
| MK | 65 | 0.621 | 0.662 | 5.67 | 0.10 | 0.080* |
| SJ | 65 | 0.676 | 0.674 | 5.59 | 0.08 | 0.013 |
| BT | 49 | 0.455 | 0.478 | 4.12 | 0.07 | 0.063 |
| KD | 62 | 0.612 | 0.588 | 5.23 | 0.42 | -0.025 |
| LD | 47 | 0.591 | 0.575 | 4.13 | 0.01 | -0.013 |
| DN | 33 | 0.334 | 0.324 | 2.91 | 0.00 | -0.012 |
| HP | 43 | 0.364 | 0.403 | 3.67 | 0.21 | 0.114* |
| YL | 36 | 0.361 | 0.430 | 3.20 | 0.00 | 0.178* |
| 平均值 Mean | 57 | 0.555 | 0.579 | 4.89 | 0.15 | - |
| DA | YS | HX | DL | WB | MK | SJ | BT | KD | LD | DN | HP | YL | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DA | - | ||||||||||||
| YS | 0.026 | - | |||||||||||
| HX | 0.017 | 0.039 | - | ||||||||||
| DL | 0.017 | 0.028 | 0.017 | - | |||||||||
| WB | 0.058 | 0.058 | 0.079 | 0.036 | - | ||||||||
| MK | 0.054 | 0.073 | 0.052 | 0.026 | 0.058 | - | |||||||
| SJ | 0.066 | 0.077 | 0.079 | 0.044 | 0.042 | 0.018 | - | ||||||
| BT | 0.203 | 0.216 | 0.194 | 0.150 | 0.192 | 0.105 | 0.100 | - | |||||
| KD | 0.087 | 0.129 | 0.103 | 0.073 | 0.093 | 0.036 | 0.080 | 0.171 | - | ||||
| LD | 0.091 | 0.118 | 0.088 | 0.067 | 0.091 | 0.019 | 0.082 | 0.167 | 0.020 | - | |||
| DN | 0.291 | 0.319 | 0.295 | 0.254 | 0.283 | 0.193 | 0.187 | 0.091 | 0.229 | 0.247 | - | ||
| HP | 0.250 | 0.274 | 0.255 | 0.202 | 0.229 | 0.141 | 0.138 | 0.050 | 0.193 | 0.196 | 0.068 | - | |
| YL | 0.216 | 0.249 | 0.227 | 0.184 | 0.204 | 0.124 | 0.120 | 0.087 | 0.158 | 0.175 | 0.076 | 0.063 | - |
Table 3 Genetic differentiation coefficient between the Betula platyphylla populations on the east side of the Qingzang Plateau
| DA | YS | HX | DL | WB | MK | SJ | BT | KD | LD | DN | HP | YL | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DA | - | ||||||||||||
| YS | 0.026 | - | |||||||||||
| HX | 0.017 | 0.039 | - | ||||||||||
| DL | 0.017 | 0.028 | 0.017 | - | |||||||||
| WB | 0.058 | 0.058 | 0.079 | 0.036 | - | ||||||||
| MK | 0.054 | 0.073 | 0.052 | 0.026 | 0.058 | - | |||||||
| SJ | 0.066 | 0.077 | 0.079 | 0.044 | 0.042 | 0.018 | - | ||||||
| BT | 0.203 | 0.216 | 0.194 | 0.150 | 0.192 | 0.105 | 0.100 | - | |||||
| KD | 0.087 | 0.129 | 0.103 | 0.073 | 0.093 | 0.036 | 0.080 | 0.171 | - | ||||
| LD | 0.091 | 0.118 | 0.088 | 0.067 | 0.091 | 0.019 | 0.082 | 0.167 | 0.020 | - | |||
| DN | 0.291 | 0.319 | 0.295 | 0.254 | 0.283 | 0.193 | 0.187 | 0.091 | 0.229 | 0.247 | - | ||
| HP | 0.250 | 0.274 | 0.255 | 0.202 | 0.229 | 0.141 | 0.138 | 0.050 | 0.193 | 0.196 | 0.068 | - | |
| YL | 0.216 | 0.249 | 0.227 | 0.184 | 0.204 | 0.124 | 0.120 | 0.087 | 0.158 | 0.175 | 0.076 | 0.063 | - |
| 变异来源 Source of variance | 变异 Variance | 变异百分比 Percentage of variance (%) |
|---|---|---|
| 种群间 Among populations | 0.477 | 13 |
| 个体间 Among individuals | 0.191 | 5 |
| 个体内 Within individuals | 3.064 | 82 |
| 总体 Total | 3.733 | 100 |
Table 4 Analysis of Molecular Variance of the Betula platyphylla populations on the east side of the Qingzang Plateau
| 变异来源 Source of variance | 变异 Variance | 变异百分比 Percentage of variance (%) |
|---|---|---|
| 种群间 Among populations | 0.477 | 13 |
| 个体间 Among individuals | 0.191 | 5 |
| 个体内 Within individuals | 3.064 | 82 |
| 总体 Total | 3.733 | 100 |
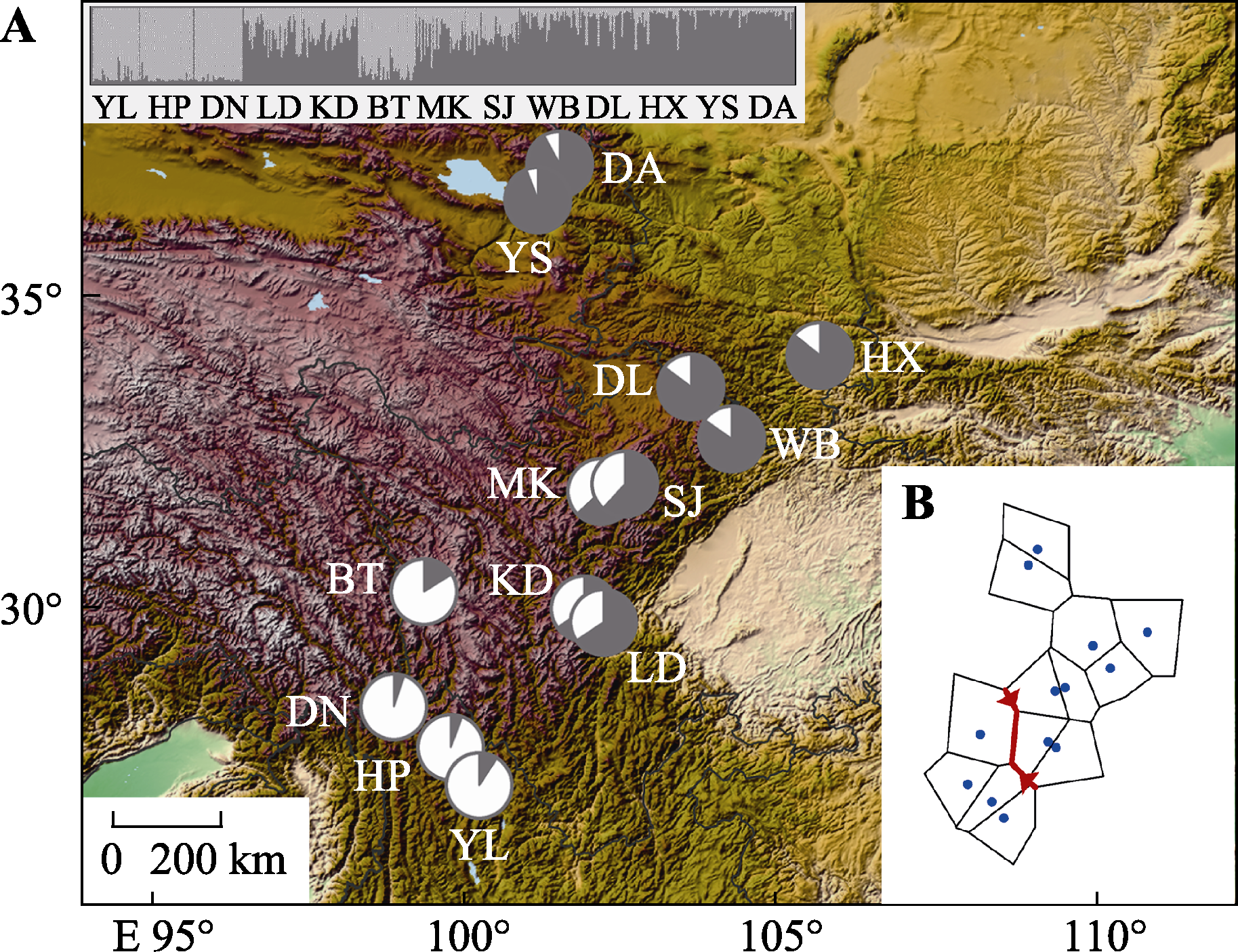
Fig. 1 Results of genetic structure analysis (A) and genetic barrier analysis (B) of the Betula platyphylla populations on the east side of the Qingzang Plateau. See Table 1 for populations.
| [1] |
Avise JC (2004). What is the field of biogeography, and where is it going? Taxon, 53, 893-898.
DOI URL |
| [2] |
Borrell JS, Wang N, Nichols RA, Buggs RJA (2018). Genetic diversity maintained among fragmented populations of a tree undergoing range contraction. Heredity, 121, 304-318.
DOI PMID |
| [3] |
Bos JAA (2001). Lateglacial and Early Holocene vegetation history of the northern Wetterau and the Amöneburger Basin (Hessen), central-west Germany. Review of Palaeobotany and Palynology, 115, 177-212.
DOI URL |
| [4] |
Bouilhol P, Jagoutz O, Hanchar JM, Dudas FO (2013). Dating the India-Eurasia collision through arc magmatic records. Earth and Planetary Science Letters, 366, 163-175.
DOI URL |
| [5] |
Cao XY, Herzschuh U, Ni J, Zhao Y, Böehmer T (2015). Spatial and temporal distributions of major tree taxa in eastern continental Asia during the last 22,000 years. The Holocene, 25, 79-91.
DOI URL |
| [6] |
Chen TY, Lou AR (2019). Phylogeography and paleodistribution models of a widespread birch (Betula platyphylla Suk.) across East Asia: multiple refugia, multidirectional expansion, and heterogeneous genetic pattern. Ecology and Evolution, 9, 7792-7807.
DOI URL |
| [7] | Chen ZD (1994). Phylogeny and phytogeography of the Betulaceae. Acta Phytotaxonomica Sinica, 32, 1-31. |
| [ 陈之端 (1994). 桦木科植物的系统发育和地理分布. 植物分类学报, 32, 1-31.] | |
| [8] |
Du FK, Hou M, Wang WT, Mao KS, Hampe A (2017). Phylogeography of Quercus aquifolioides provides novel insights into the Neogene history of a major global hotspot of plant diversity in south-west China. Journal of Biogeography, 44, 294-307.
DOI URL |
| [9] |
Evanno G, Regnaut S, Goudet J (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology, 14, 2611-2620.
PMID |
| [10] |
Forest F, Savolainen V, Chase MW, Lupia R, Bruneau A, Crane PR (2005). Teasing apart molecular-versus fossil- based error estimates when dating phylogenetic trees: a case study in the birch family (Betulaceae). Systematic Botany, 30, 118-133.
DOI URL |
| [11] | Goudet J (2001). FSTAT, A program to estimate and test gene diversities and fixation indices, version 2.9.3. [2021-10-5]. http://www2.unil.ch/popgen/softwares/fstat.html. |
| [12] | Guan WB (1998). Vegetation ecology on communities dominated by Betula platyphylla in northeast of China distribution of communities dominated by Betula platyphylla. Journal of Beijing Forestry University, 20(4), 104-109. |
| [ 关文彬 (1998). 中国东北地区白桦林植被生态学的研究——桦属植物与中国白桦林的地理分布. 北京林业大学学报, 20(4), 104-109.] | |
| [13] | He K, Jiang XL (2014). Sky islands of southwest China: I. An overview of phylogeographic patterns. Chinese Science Bulletin, 59, 1055-1068. |
| [ 何锴, 蒋学龙 (2014). 中国西南地区的“天空之岛”: I系统地理学研究概述. 科学通报, 59, 1055-1068.] | |
| [14] |
Herzschuh U, Kürschner H, Mischke S (2006). Temperature variability and vertical vegetation belt shifts during the last -50,000 yr in the Qilian Mountains (NE margin of the Tibetan Plateau, China). Quaternary Research, 66, 133-146.
DOI URL |
| [15] |
Hewitt G (2000). The genetic legacy of the Quaternary ice ages. Nature, 405, 907-913.
DOI URL |
| [16] |
Hu YN, Zhao L, Buggs RJA, Zhang XM, Li J, Wang N (2019). Population structure of Betula albosinensis and Betula platyphylla: evidence for hybridization and a cryptic lineage. Annals of Botany, 123, 1179-1189.
DOI URL |
| [17] |
Huang JH, Chen B, Liu CR, Lai JS, Zhang JL, Ma KP (2012). Identifying hotspots of endemic woody seed plant diversity in China. Diversity and Distributions, 18, 673-688.
DOI URL |
| [18] | Jadwiszczak K (2012). What can molecular markers tell us about the glacial and postglacial histories of European birches? Silva Fennica, 46, 733-745. |
| [19] |
Jagoutz O, Royden L, Holt AF, Becker TW (2015). Anomalously fast convergence of India and Eurasia caused by double subduction. Nature Geoscience, 8, 475-478.
DOI |
| [20] | Jiang JM (1990). The study of the geographical distribution of the Betula in China. Forest Research, 3, 55-62. |
| [ 姜景民 (1990). 中国桦木属植物地理分布的研究. 林业科学研究, 3, 55-62.] | |
| [21] |
Jiang K, Gao H, Chen XY (2018). Clonal diversity and genetic structure of Enhalus acoroides populations along Hainan Island, China. Chinese Journal of Applied Ecology, 29, 397-402.
DOI PMID |
|
[ 蒋凯, 高辉, 陈小勇 (2018). 海南岛海菖蒲种群克隆多样性和遗传结构. 应用生态学报, 29, 397-402.]
PMID |
|
| [22] |
Kalinowski ST (2005). Hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Molecular Ecology Notes, 5, 187-189.
DOI URL |
| [23] |
Kang M, Wang J, Huang HW (2008). Demographic bottlenecks and low gene flow in remnant populations of the critically endangered Berchemiella wilsonii var. pubipetiolata (Rhamnaceae) inferred from microsatellite markers. Conservation Genetics, 9, 191-199.
DOI URL |
| [24] |
Keppel G, van Niel KP, Wardell-Johnson GW, Yates CJ, Byrne M, Mucina L, Schut AGT, Hopper SD, Franklin SE (2012). Refugia: identifying and understanding safe havens for biodiversity under climate change. Global Ecology and Biogeography, 21, 393-404.
DOI URL |
| [25] |
Kulju KKM, Pekkinen M, Varvio S (2004). Twenty-three microsatellite primer pairs for Betula pendula (Betulaceae). Molecular Ecology Notes, 4, 471-473.
DOI URL |
| [26] |
Li ZH, Zou JB, Mao KS, Lin K, Li HP, Liu JQ, Källman T, Lascoux M (2012). Population genetic evidence for complex evolutionary histories of four high altitude Juniper species in the Qinghai-Tibetan Plateau. Evolution, 66, 831-845.
DOI URL |
| [27] |
Lind-Riehl J, Gailing O (2015). Fine-scale spatial genetic structure of two red oak species, Quercus rubra and Quercus ellipsoidalis. Plant Systematics and Evolution, 301, 1601-1612.
DOI URL |
| [28] | Liu GW, Li JG (2016). Miocene fossil floral horizons in the oiyug basin, southern central Tibet, and related stratigraphic problems. Journal of Stratigraphy, 40, 92-99. |
| [ 刘耕武, 李建国 (2016). 西藏南木林乌郁盆地中新世植物化石层位及相关地层问题. 地层学杂志, 40, 92-99.] | |
| [29] |
Liu J, Luo YH, Li DZ, Gao LM (2017). Evolution and maintenance mechanisms of plant diversity in the Qinghai-Tibet Plateau and adjacent regions: retrospect and prospect. Biodiversity Science, 25, 163-174.
DOI |
|
[ 刘杰, 罗亚皇, 李德铢, 高连明 (2017). 青藏高原及毗邻区植物多样性演化与维持机制: 进展及展望. 生物多样性, 25, 163-174.]
DOI |
|
| [30] |
Liu J, Möller M, Provan J, Gao LM, Poudel RC, Li DZ (2013). Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot. New Phytologist, 199, 1093-1108.
DOI URL |
| [31] |
Liu JQ, Sun YS, Ge XJ, Gao LM, Qiu YX (2012). Phylogeographic studies of plants in China: advances in the past and directions in the future. Journal of Systematics and Evolution, 50, 267-275.
DOI URL |
| [32] |
López-Pujol J, Zhang FM, Sun HQ, Ying TS, Ge S (2011a). Centres of plant endemism in China: places for survival or for speciation? Journal of Biogeography, 38, 1267-1280.
DOI URL |
| [33] |
López-Pujol J, Zhang FM, Sun HQ, Ying TS, Ge S (2011b). Mountains of Southern China as “plant museums” and “plant cradles”: evolutionary and conservation insights. Mountain Research and Development, 31, 261-269.
DOI URL |
| [34] |
Manni F, Guérard E, Heyer E (2004). Geographic patterns of (genetic, morphologic, linguistic) variation: How barriers can be detected by using Monmonier’s algorithm. Human Biology, 76, 173-190.
DOI URL |
| [35] |
Markova AK, Simakova AN, Puzachenko AY (2009). Ecosystems of Eastern Europe at the time of maximum cooling of the Valdai glaciation (24-18 kyr BP) inferred from data on plant communities and mammal assemblages. Quaternary International, 201, 53-59.
DOI URL |
| [36] | Mittermeier RA, Turner WR, Larsen FW, Brooks TM, Gascon C (2011). Global biodiversity conservation: the critical role of hotspots//Zachos F, Habel J. Biodiversity Hotspots. Springer, Berlin. 3-22. |
| [37] |
Peakall R, Smouse PE (2006). Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6, 288-295.
DOI URL |
| [38] |
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945-959.
DOI PMID |
| [39] |
Qiu YX, Fu CX, Comes HP (2011). Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the worldʼs most diverse temperate flora. Molecular Phylogenetics and Evolution, 59, 225-244.
DOI URL |
| [40] |
Shen TT, Ran JH, Wang XQ (2019). Phylogenomics disentangles the evolutionary history of spruces (Picea) in the Qinghai- Tibetan Plateau: implications for the design of population genetic studies and species delimitation of conifers. Molecular Phylogenetics and Evolution, 141, 106612. DOI: 10.1016/j.ympev.2019.106612.
DOI URL |
| [41] | Shi FC, Li JW, Koike T, Nie SQ (2001). Resources of the white birch (Betula platyphylla) for sap production and its ecological characteristics in Northeast China. Eurasian Journal of Forest Research, 2, 31-38. |
| [42] |
Sofiev M, Siljamo P, Ranta H, Rantio-Lehtimäki A (2006). Towards numerical forecasting of long-range air transport of birch pollen: theoretical considerations and a feasibility study. International Journal of Biometeorology, 50, 392-402.
PMID |
| [43] | Su ZH, Li WJ, Zhuo L, Jiang XL (2017). Analyses on genetic variation and genetic structure of populations of endangered plant Helianthemum songaricum in Xinjiang. Journal of Plant Resources and Environment, 26(4), 67-73. |
| [ 苏志豪, 李文军, 卓立, 姜小龙 (2017). 新疆濒危植物半日花居群的遗传变异及遗传结构分析. 植物资源与环境学报, 26(4), 67-73.] | |
| [44] |
Tang ZY, Wang ZH, Fang JY (2009). Historical hypothesis in explaining spatial patterns of species richness. Biodiversity Science, 17, 635-643.
DOI URL |
|
[ 唐志尧, 王志恒, 方精云 (2009). 生物多样性分布格局的地史成因假说. 生物多样性, 17, 635-643.]
DOI |
|
| [45] |
Tsuda Y, Semerikov V, Sebastiani F, Vendramin GG, Lascoux M (2017). Multispecies genetic structure and hybridization in the Betula genus across Eurasia. Molecular Ecology, 26, 589-605.
DOI URL |
| [46] |
Tsuda Y, Ueno S, Ranta J, Salminen K, Ide Y, Shinohara K, Tsumura Y (2009). Development of 11 EST-SSRs for Japanese white birch, Betula platyphylla var. japonica and their transferability to related species. Conservation Genetics, 10, 1385-1388.
DOI URL |
| [47] |
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004). Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes, 4, 535-538.
DOI URL |
| [48] |
Wang N, Borrell JS, Bodles WJA, Kuttapitiya A, Nichols RA, Buggs RJA (2014). Molecular footprints of the Holocene retreat of dwarf birch in Britain. Molecular Ecology, 23, 2771-2782.
DOI URL |
| [49] |
Willis KJ, Rudner E, Sümegi P (2000). The full-glacial forests of central and southeastern Europe. Quaternary Research, 53, 203-213.
DOI URL |
| [50] |
Willis KJ, van Andel TH, (2004). Trees or no trees? The environments of central and eastern Europe during the Last Glaciation. Quaternary Science Reviews, 23, 2369-2387.
DOI URL |
| [51] |
Wu B, Lian C, Hogetsu T (2002). Development of microsatellite markers in white birch (Betula platyphylla var. japonica). Molecular Ecology Notes, 2, 413-415.
DOI URL |
| [52] |
Yan YJ, Yang X, Tang ZY (2013). Patterns of species diversity and phylogenetic structure of vascular plants on the Qinghai-Tibetan Plateau. Ecology and Evolution, 3, 4584-4595.
DOI URL |
| [53] | Yang WB, Lan SB, Li HY (2012). A review of the geographical distribution and utilization of the Betula. Journal of Heilongjiang Vocational Institute of Ecological Engineering, 25(4), 18-19. |
| [ 杨万波, 兰士波, 李红艳 (2012). 桦木属植物地理分布及利用价值述评. 黑龙江生态工程职业学院学报, 25(4), 18-19.] | |
| [54] | Yin DX (2013). Phylogeography and Population Structure of Betula platyphylla. Master degree dissertation, Northeast Normal University, Changchun. |
| [ 尹东旭 (2013). 白桦的亲缘地理与遗传结构研究. 硕士学位论文, 东北师范大学, 长春.] | |
| [55] |
Young A, Boyle T, Brown T (1996). The population genetic consequences of habitat fragmentation for plants. Trends in Ecology & Evolution, 11, 413-418.
DOI URL |
| [56] | Yu HB, Zhang YL (2013). Advances in phylogeography of alpine plants in the Tibetan Plateau and adjacent regions. Acta Botanica Boreali-Occidentalia Sinica, 33, 1268-1278. |
| [ 于海彬, 张镱锂 (2013). 青藏高原及其周边地区高山植物谱系地理学研究进展. 西北植物学报, 33, 1268-1278.] | |
| [57] | Zyryanova O, Terazawa M, Koike T, Zyryanov VI (2010). White birch trees as resource species of Russia: their distribution, ecophysiological features, multiple utilizations. Eurasian Journal of Forest Research, 13, 25-40. |
| [1] | XU Jin-Shi,CHAI Yong-Fu,LIU Xiao,YUE Ming,GUO Yao-Xin,KANG Mu-Yi,LIU Quan-Ru,ZHENG Cheng-Yang,JI Cheng-Jun,YAN Ming,ZHANG Feng,GAO Xian-Ming,WANG Ren-Qing,SHI Fu-Chen,ZHANG Qin-Di,WANG Mao. Community assembly, diversity patterns and distributions of broad-leaved forests in North China [J]. Chin J Plant Ecol, 2019, 43(9): 732-741. |
| [2] | QIN Hao, ZHANG Yin-Bo, DONG Gang, ZHANG Feng. Altitudinal patterns of taxonomic, phylogenetic and functional diversity of forest communities in Mount Guandi, Shanxi, China [J]. Chin J Plant Ecol, 2019, 43(9): 762-773. |
| [3] | ZHANG Xin-Xin, WANG Xi, HU Ying, ZHOU Wei, CHEN Xiao-Yang, HU Xin-Sheng. Advances in the study of population genetic diversity at plant species’ margins [J]. Chin J Plant Ecol, 2019, 43(5): 383-395. |
| [4] | CHENG Yi-Kang, ZHANG Hui, WANG Xu, LONG Wen-Xing, LI Chao, FANG Yan-Shan, FU Ming-Qi, ZHU Kong-Xin. Effects of functional diversity and phylogenetic diversity on the tropical cloud forest community assembly [J]. Chin J Plant Ecol, 2019, 43(3): 217-226. |
| [5] | ZHANG Li-Wen, HAN Guang-Xuan. A review on the relationships between plant genetic diversity and ecosystem functioning [J]. Chin J Plan Ecolo, 2018, 42(10): 977-989. |
| [6] | Xi-Xi WANG, Wen-Xing LONG, Xiao-Bo YANG, Meng-Hui XIONG, Yong KANG, Jin HUANG, Xu WANG, Xiao-Jiang HONG, Zhao-Li ZHOU, Yong-Quan LU, Jing FANG, Shi-Xing LI. Patterns of plant diversity within and among three tropical cloud forest communities in Hainan Island [J]. Chin J Plant Ecol, 2016, 40(5): 469-479. |
| [7] | WANG Jin-Nan,CHEN Jin-Fu,Chen Wu-Sheng,Zhou Xin-Yang,XU Dong,LI Ji-Hong,QI Xiao. Population genetic diversity of wild Lycium ruthenicum in Qaidam inferred from AFLP markers [J]. Chin J Plan Ecolo, 2015, 39(10): 1003-1011. |
| [8] | LIU Jun, JIANG Jing-Min, ZOU Jun, XU Jin-Liang, SHEN Han, DIAO Song-Feng. Genetic diversity of central and peripheral populations of Toona ciliata var. pubescens, an endangered tree species endemic to China [J]. Chin J Plant Ecol, 2013, 37(1): 52-60. |
| [9] | ZHANG Wei, LUO Jian-Xun, GU Yun-Jie, HU Ting-Xing. Allozyme variation of genetic diversity in natural populations of Jatropha curcas germplasm from different areas in southwest China [J]. Chin J Plant Ecol, 2011, 35(3): 330-336. |
| [10] | WEI Yuan, WANG Shi-Jie, LIU Xiu-Ming, HUANG Tian-Zhi. Genetic diversity of arbuscular mycorrhizal fungi in karst microhabitats of Guizhou Province, China [J]. Chin J Plant Ecol, 2011, 35(10): 1083-1090. |
| [11] | ZHANG Yun-Hong, HOU Yan, LOU An-Ru. Population genetic diversity of Rhodiola dumulosa in Northern China inferred from AFLP makers [J]. Chin J Plant Ecol, 2010, 34(9): 1084-1094. |
| [12] | CHEN Ying. DETECTING EFFECT OF PHYLOGENETIC DIVERSITY ON SEEDLING MORTALITY IN AN EVERGREEN BROAD-LEAVED FOREST IN CHINA [J]. Chin J Plant Ecol, 2009, 33(6): 1084-1089. |
| [13] | LIU Wei, WANG Xi, GAN You-Min, HUANG Lin-Kai, XIE Wen-Gang, MIAO Jia-Min. GENETIC DIVERSITY OF KOBRESIA PYGMAEA POPULATIONS ALONG A GRAZING GRADIENT [J]. Chin J Plant Ecol, 2009, 33(5): 966-973. |
| [14] | CHEN Liang-Hua, HU Ting-Xing, ZHANG Fan, LI Guo-He. GENETIC DIVERSITIES OF FOUR JUGLANSPOPULATIONS REVEALED BY AFLP IN SICHUAN PROVINCE, CHINA [J]. Chin J Plant Ecol, 2008, 32(6): 1362-1372. |
| [15] | ZHOU Hui-Ping, CHEN Jin, ZHANG Shou-Zhou. GENETIC AND CLONAL DIVERSITY OF GLOBBA RACEMOSA, A HERB WITH A MIXED REPRODUCTIVE MODE [J]. Chin J Plant Ecol, 2008, 32(4): 751-759. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Chinese Journal of Plant Ecology
Tel: 010-62836134, 62836138, E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn