

Chin J Plant Ecol ›› 2024, Vol. 48 ›› Issue (10): 1326-1335.DOI: 10.17521/cjpe.2023.0360 cstr: 32100.14.cjpe.2023.0360
• Research Articles • Previous Articles Next Articles
MA Jia-Zheng1, CHEN Yu-Ting1, MA Song-Mei1,*( )(
)( ), ZHANG Dan2, HE Ling-Yun1
), ZHANG Dan2, HE Ling-Yun1
Received:2023-12-04
Accepted:2024-05-22
Online:2024-10-20
Published:2024-12-03
Contact:
MA Song-Mei
Supported by:MA Jia-Zheng, CHEN Yu-Ting, MA Song-Mei, ZHANG Dan, HE Ling-Yun. Genetic pattern and diffusion path simulation of Haloxylon persicum in Xinjiang based on multi-source data[J]. Chin J Plant Ecol, 2024, 48(10): 1326-1335.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.plant-ecology.com/EN/10.17521/cjpe.2023.0360
| 序列 Sequence | 单倍型多样性 Haplotype diversity | 核苷酸多样性 Nucleotide diversity | 总遗传多样性 Total genetic diversity | 种群内平均遗传多样性 Average genetic diversity within populations | GST | NST |
|---|---|---|---|---|---|---|
| cpDNA | 0.749 | 0.000 95 | 0.862* | 0.155 | 0.820 | 0.832 |
| nrDNA | 0.600 | 0.001 42 | 0.777* | 0.217 | 0.764 | 0.723 |
Table 1 Genetic diversity and coefficient of genetic differentiation for Haloxylon persicum in Xinjiang
| 序列 Sequence | 单倍型多样性 Haplotype diversity | 核苷酸多样性 Nucleotide diversity | 总遗传多样性 Total genetic diversity | 种群内平均遗传多样性 Average genetic diversity within populations | GST | NST |
|---|---|---|---|---|---|---|
| cpDNA | 0.749 | 0.000 95 | 0.862* | 0.155 | 0.820 | 0.832 |
| nrDNA | 0.600 | 0.001 42 | 0.777* | 0.217 | 0.764 | 0.723 |
| 变异来源 Source of variation | 自由度 df | 平方和 Sum of squares | 变异组成 Variance components | 变异所占比例 Percentage of variation (%) | 固定指数 Fixation index (FST) | |
|---|---|---|---|---|---|---|
| cpDNA | 种群间 Among populations | 12 | 6.750 | 0.69 | 80.65 | 0.81 |
| 种群内 Within populations | 94 | 1.333 | 0.17 | 19.35 | ||
| 总变异 Total variation | 106 | 8.083 | 0.86 | |||
| nrDNA | 种群间 Among populations | 12 | 5.417 | 0.55 | 76.62 | 0.76 |
| 种群内 Within populations | 94 | 1.333 | 0.17 | 23.38 | ||
| 总变异 Total variation | 106 | 6.750 | 0.71 |
Table 2 Analysis of molecular variance (AMOVA) for populations of Haloxylon persicum in Xinjiang
| 变异来源 Source of variation | 自由度 df | 平方和 Sum of squares | 变异组成 Variance components | 变异所占比例 Percentage of variation (%) | 固定指数 Fixation index (FST) | |
|---|---|---|---|---|---|---|
| cpDNA | 种群间 Among populations | 12 | 6.750 | 0.69 | 80.65 | 0.81 |
| 种群内 Within populations | 94 | 1.333 | 0.17 | 19.35 | ||
| 总变异 Total variation | 106 | 8.083 | 0.86 | |||
| nrDNA | 种群间 Among populations | 12 | 5.417 | 0.55 | 76.62 | 0.76 |
| 种群内 Within populations | 94 | 1.333 | 0.17 | 23.38 | ||
| 总变异 Total variation | 106 | 6.750 | 0.71 |
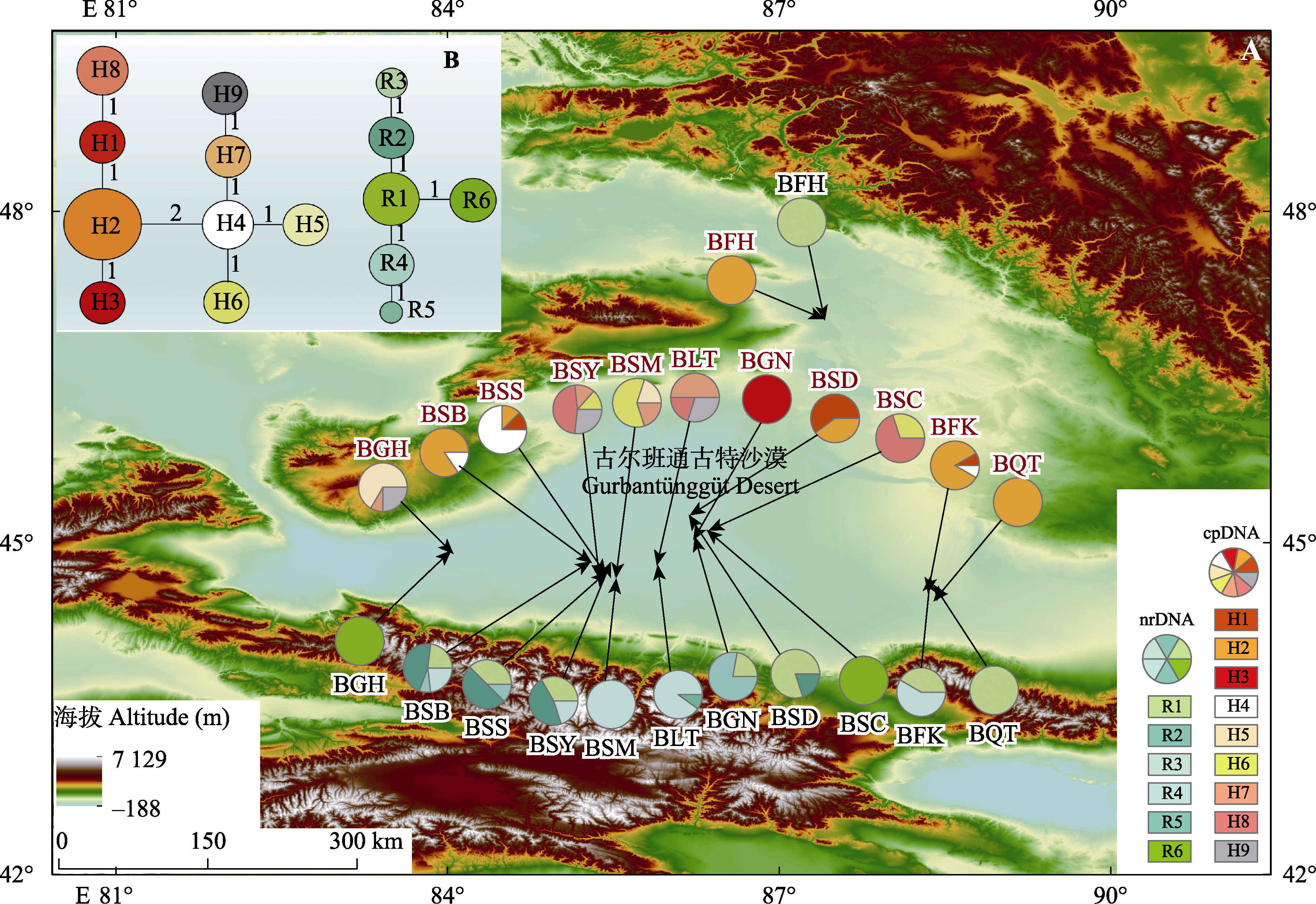
Fig. 1 Sampling localities, geographic distribution (A) and the haplotype network (B) of 9 chloroplast DNA (cpDNA) haplotypes (labelled as H1-H9) and 6 nuclear ribosomal DNA (nrDNA) haplotypes (labelled as R1-R6), identified from 12 populations of Haloxylon persicum. BFH, gold coast of Fuhai County, Altay Region; BFK, Fukang City; BGH, Ganjiahu Saxoul Nature Reserve; BGN, southern edge of Gurbantünggüt Desert; BLT, Kuytun County, Shawan City; BQT, Beishawo of Qitai County; BSB, Shamenzi Town of Shihezi Corps 134th; BSC, Mosuowan Reclamation Area of Shihezi Corps 149th; BSD, Shihezi Corps 150th; BSM, desert of northern Shihezi Corps 147th; BSS, Anjihai Town of Shihezi Corps 134th; BSY, Xiayedi Town of Shihezi Corps 134th.
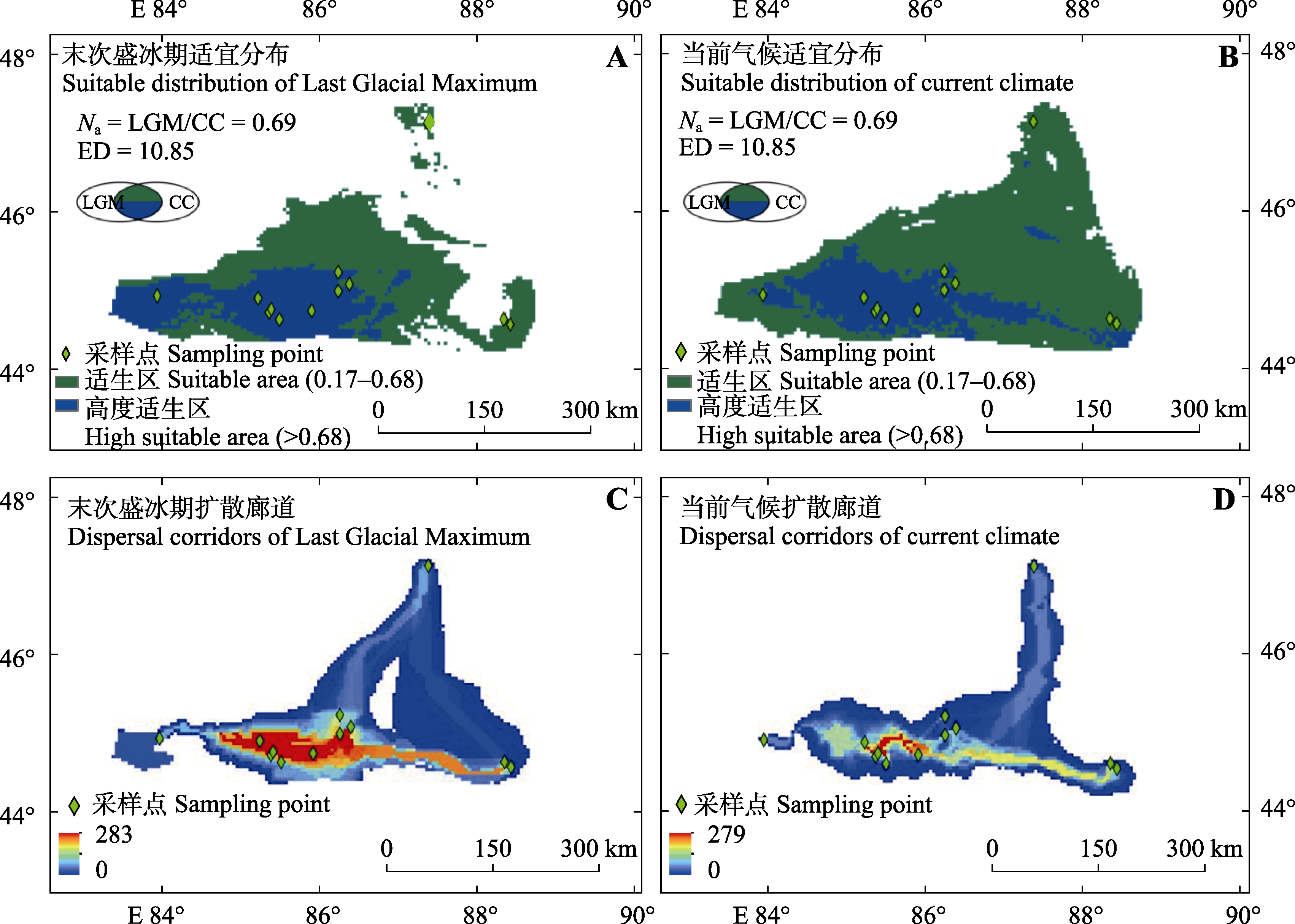
Fig. 2 Suitable distribution (A, B) and dispersal corridors (C, D) for Haloxylon persicum in Gurbantünggüt Desert in Last Glacial Maximum and current climate. The colors in figures C and D from blue to red represent the possibility of potential dispersal paths from low to high. CC, current climate; ED, edge density; LGM, Last Glacial Maximum; Na, habitat distribution area ratio.
| 序列 Sequence | Fu’s Fs | Tajima’s D | 离差平方和 Sum of squared deviation | 粗糙度指数 Raggedness index |
|---|---|---|---|---|
| cpDNA | 1.259 | 1.264 | 0.094* | 0.273* |
| nrDNA | 2.024 | 1.665 | 0.020* | 0.211* |
Table 3 Results of neutrality tests for all populations of Haloxylon persicum based on the cpDNA and nrDNA dataset
| 序列 Sequence | Fu’s Fs | Tajima’s D | 离差平方和 Sum of squared deviation | 粗糙度指数 Raggedness index |
|---|---|---|---|---|
| cpDNA | 1.259 | 1.264 | 0.094* | 0.273* |
| nrDNA | 2.024 | 1.665 | 0.020* | 0.211* |
| [1] | Abbott RJ, Comes HP (2004). Evolution in the Arctic: a phylogeographic analysis of the circumarctic plant (purple saxifrage). New Phytologist, 161, 211-224. |
| [2] | An ZY, Tang H, Li WR (2018). Genetic diversity analysis of Paeonia rockii cultivar based on EST-SSR. Molecular Plant Breeding, 16, 6744-6752. |
| [ 安宗燕, 唐红, 李婉茹 (2018). 基于EST-SSR的紫斑牡丹品种遗传多样性分析. 分子植物育种, 16, 6744-6752.] | |
| [3] | Chen YT, Ma SM, Zhang D, Wei B, Huang G, Zhang YL, Ge BW (2022). Diversification and historical demography of Haloxylon ammodendron in relation to Pleistocene climatic oscillations in northwestern China. PeerJ, 10, e14476. DOI: 10.7717/peerj.14476. |
| [4] | Chen YT, Ma SM, Zhang D, Zhang L, Wang CC (2024). Diversity pattern and formation mechanism of sympatric Haloxylon ammodendron and Haloxylon persicum in Xinjiang, China. Chinese Journal of Plant Ecology, 48, 56-67. |
|
[ 陈雨婷, 马松梅, 张丹, 张林, 王春成 (2024). 新疆同域分布梭梭和白梭梭多样性格局及其形成机制. 植物生态学报, 48, 56-67.]
DOI |
|
| [5] | Dong GR, Chen HZ, Wang GY, Li XZ, Shao YJ, Jin J (1995). Evolution and climate change of desert and sandy land in Northern China since 150 ka. Science in China (Series B), 25, 1303-1312. |
| [ 董光荣, 陈惠忠, 王贵勇, 李孝泽, 邵亚军, 金炯 (1995). 150 ka以来中国北方沙漠、沙地演化和气候变化. 中国科学(B辑), 25, 1303-1312.] | |
| [6] | Excoffier L, Laval G, Schneider S (2007). Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online, 1, 47-50. |
| [7] | Feng XL, Liu R, Ma J, Xu Z, Wang YG, Kong L (2021). Photosynthetic characteristics and influencing factors of Haloxylon persicum stems (different diameter classes) in Gurbantonggut Desert. Acta Ecologica Sinica, 41, 9784-9795. |
| [ 冯晓龙, 刘冉, 马健, 徐柱, 王玉刚, 孔璐 (2021). 古尔班通古特沙漠白梭梭枝干光合及其影响因素. 生态学报, 41, 9784-9795.] | |
| [8] | Group TAP (2003). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Botanical Journal of the Linnean Society, 141, 399-436. |
| [9] | Group TAP (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society, 181, 1-20. |
| [10] |
Harpending HC (1994). Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Human Biology, 66, 591-600.
PMID |
| [11] | Hewitt GM (2004). Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society of Series B: Biological Sciences, 359, 183-195. |
| [12] | Jiang XL, Xu GB, Deng M (2019). Spatial genetic patterns and distribution dynamics of the rare oak Quercus chungii: implications for biodiversity conservation in southeast China. Forests, 10, 821. DOI: 10.3390/f10090821. |
| [13] | Jin K, Lu Y, Zhou HM, Zhang QZ, Hu Y, Wan D, Yan JM (2022). Research progress on the hydrology in the Gurbantunggut Desert. Journal of China Hydrology, 42(1), 1-10. |
| [ 金可, 卢阳, 周火明, 张乾柱, 胡月, 万丹, 闫建梅 (2022). 古尔班通古特沙漠水文研究进展. 水文, 42(1), 1-10.] | |
| [14] | Lang XP, Fan RY, Li QF (2023). Analysis of potential suitable areas of Allium mongolicum in northern China. Acta Agrestia Sinica, 31, 3525-3534. |
|
[ 郎显鹏, 樊如月, 李青丰 (2023). 中国北方地区蒙古韭潜在适生区分析. 草地学报, 31, 3525-3534.]
DOI |
|
| [15] | Li X, Gao GL, Sun GL, Shi HB, Zhao FF, Ma L (2021). Potential suitable areas of Haloxylon ammodendron and Haloxylon persicum in Xinjiang based on MaxEnt. Journal of West China Forestry Science, 50(1), 145-152. |
| [ 李雪, 高广磊, 孙桂丽, 史浩伯, 赵芳芳, 马龙 (2021). 基于MaxEnt预测梭梭和白梭梭在新疆的潜在适生区. 西部林业科学, 50(1), 145-152.] | |
| [16] | Ma SM, Nie YB, Jiang XL, Xu Z, Ji WQ (2019). Genetic structure of the endangered, relict shrub Amygdalus mongolica (Rosaceae) in arid Northwest China. Australian Journal of Botany, 67, 128-139. |
| [17] | Mao ZM, Zhang DM (1994). The conspectus of ephemeral flora in northern Xinjiang. Arid Zone Research, 11(3), 1-26. |
| [ 毛祖美, 张佃民 (1994). 新疆北部早春短命植物区系纲要. 干旱区研究, 11(3), 1-26.] | |
| [18] | “Natural Environmental Evolution” Research Group (2003). Evolution and development trend of natural environment in northwest China. China Water Resources, 9, 33-36. |
| [ “自然环境演变”课题组 (2003). 西北地区自然环境演变及其发展趋势. 中国水利, 9, 33-36.] | |
| [19] | Reed DH, Frankham R (2003). Correlation between fitness and genetic diversity. Conservation Biology, 17, 230-237. |
| [20] |
Richardson JE, Pennington RT, Pennington TD, Hollingsworth PM (2001). Rapid diversification of a species-rich genus of neotropical rain forest trees. Science, 293, 2242-2245.
DOI PMID |
| [21] |
Shaw J, Lickey EB, Schilling EE, Small RL (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. American Journal of Botany, 94, 275-288.
DOI PMID |
| [22] | Sun FF, Nie YB, Ma SM, Wei B, Ji WQ (2019). Species differentiation of Haloxylon ammodendron and Haloxylon persicum based on ITS and cpDNA sequences. Scientia Silvae Sinicae, 55(3), 43-53. |
| [ 孙芳芳, 聂迎彬, 马松梅, 魏博, 吉万全 (2019). 基于ITS和cpDNA序列的梭梭和白梭梭物种分化. 林业科学, 55(3), 43-53.] | |
| [23] | The Editorial Committee of Vegetation of China (1980). Vegetation of China. Science Press, Beijing. 956-979. |
| [ 中国植被编辑委员会 (1980). 中国植被. 科学出版社, 北京. 956-979.] | |
| [24] | Wang CC, Ma SM, Zhang D, Wang SM (2020). Spatial genetic structure of Lycium ruthenicum in the Qaidam Basin. Chinese Journal of Plant Ecology, 44, 661-668. |
|
[ 王春成, 马松梅, 张丹, 王绍明 (2020). 柴达木野生黑果枸杞的空间遗传结构. 植物生态学报, 44, 661-668.]
DOI |
|
| [25] | Wang CL, Guo QS, Tan DY, Shi ZM, Ma C (2005). Haloxylon ammodendron community patterns in different habitats along southeastern edge of Zhunger Basin. Chinese Journal of Applied Ecology, 16, 1224-1229. |
| [ 王春玲, 郭泉水, 谭德远, 史作民, 马超 (2005). 准噶尔盆地东南缘不同生境条件下梭梭群落结构特征研究. 应用生态学报, 16, 1224-1229.] | |
| [26] | Wang Q, Zhang ML, Yin LK (2016). Phylogeographic structure of a Tethyan relict Capparis spinosa (Capparaceae) traces Pleistocene geologic and climatic changes in the western Himalayas, Tianshan Mountains, and adjacent desert regions. BioMed Research International, 2016, 5792708. DOI: 10.1155/2016/5792708. |
| [27] | Wang YT, Zhang DH, Zhang ZS (2022). Spatial distribution and interspecific correlation of Haloxylon persicum and H. ammodendron on fixed dunes of the Gurbantunggut Desert, China. Biodiversity Science, 30, 24-35. |
| [ 王雅婷, 张定海, 张志山 (2022). 古尔班通古特沙漠固定沙丘上白梭梭和梭梭的空间分布及种间关联性. 生物多样性, 30, 24-35.] | |
| [28] | Wei B, Ma SM, Song J, He LY, Li XC (2019). Prediction of the potential distribution and ecological suitability of Fritillaria walujewii. Acta Ecologica Sinica, 39, 228-234. |
| [ 魏博, 马松梅, 宋佳, 贺凌云, 李晓辰 (2019). 新疆贝母潜在分布区域及生态适宜性预测. 生态学报, 39, 228-234.] | |
| [29] | Wei Y, Yin LK, Yan C (2005). Study on the flowering and pollination characteristics of Haloxylon persicum. Arid Zone Research, 22, 85-89. |
| [ 魏岩, 尹林克, 严成 (2005). 白梭梭开花及风媒传粉特点. 干旱区研究, 22, 85-89.] | |
| [30] | White TJ, Bruns T, Lee S, Taylor J (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols, 18, 315-322. |
| [31] |
Xie XH, Li ZZ, Jin JH, Liu R, Zou XJ, Ma YQ (2022). Preliminary study on sedimentary structure and development model of vegetated linear dune in the southeastern Gurbantunggut Desert. Journal of Desert Research, 42(3), 74-84.
DOI |
|
[ 解锡豪, 李志忠, 靳建辉, 刘瑞, 邹晓君, 马运强 (2022). 古尔班通古特沙漠东南部植被线形沙丘内部构造及发育模式. 中国沙漠, 42(3), 74-84.]
DOI |
|
| [32] | Yang SF (2021). Combining the Responses of Habitat Suitability and Connectivity to Climate Change for Sika Deer (Cervus nippon) in Shennongjia Area, China. Mater degree dissertation, Central China Normal University, Shanghai. |
| [ 杨绍法 (2021). 气候变化对神农架梅花鹿生境及其连通性的影响. 硕士学位论文, 华中师范大学, 上海.] | |
| [33] | Yu HB, Zhang YL, Li SC, Qi W, Hu ZJ (2014). Predicting the dispersal routes of alpine plant Pedicularis longiflora (Orobanchaceae) based on GIS and species distribution models. Chinese Journal of Applied Ecology, 25, 1669-1673. |
| [ 于海彬, 张镱锂, 李士成, 祁威, 胡忠俊 (2014). 基于GIS和物种分布模型的高山植物长花马先蒿迁移路线模拟. 应用生态学报, 25, 1669-1673.] | |
| [34] | Zhang LY (2002). Haloxylon ammodendron and Haloxylon persicum in Xinjiang desert (Part 1). Journal of Botany, 4, 4-6. |
| [ 张立运 (2002). 新疆荒漠中的梭梭和白梭(上). 植物杂志, 4, 4-6.] | |
| [35] | Zhang P (2006). Studies on Genetic Diversity of Haloxylon in Xinjiang with ISSR. Mater degree dissertation, Xinjiang Agricultural University, Ürümqi. |
| [ 张萍 (2006). 利用ISSR分子标记对新疆梭梭属植物遗传多样性的研究. 硕士学位论文, 新疆农业大学, 乌鲁木齐.] | |
| [36] | Zhang P, Dong YZ, Wei Y, Hu CZ (2006). Analysis of genetic diversity of Haloxylon persicum (Chenopodiaceae) in Xinjiang by ISSR. Acta Botanica Yunnanica, 28, 359-362. |
| [ 张萍, 董玉芝, 魏岩, 胡成志 (2006). 利用ISSR标记对新疆白梭梭居群的遗传多样性分析. 云南植物研究, 28, 359-362.] | |
| [37] | Zhang Z (2021). Population Restoration and Habitat Protection of Red Deer (Cervus canadensis) in Chifeng. Mater degree dissertation, Beijing Forestry University, Beijing. |
| [ 张沼(2021). 赤峰市马鹿(Cervus canadensis)种群恢复和栖息地保护研究. 硕士学位论文, 北京林业大学, 北京.] | |
| [38] | Zhu BQ, Yu JJ, Qin XG, Liu ZT, Xiong HG (2013). Formation and evolution of sandy deserts in Xinjiang: the palaeo- environmental evidences. Acta Geographica Sinica, 68, 661-679. |
| [ 朱秉启, 于静洁, 秦晓光, 刘子亭, 熊黑钢 (2013). 新疆地区沙漠形成与演化的古环境证据. 地理学报, 68, 661-679.] |
| [1] | LIAO Su-Hui, NI Long-Kang, QIN Jia-Shuang, TAN Yu, GU Da-Xing. Hydraulic regulation strategies of karst forest species exhibit variation across different successional stages in the mid-subtropical zone [J]. Chin J Plant Ecol, 2024, 48(9): 1223-1231. |
| [2] | CAI Hui-Ying, LIANG Ya-Tao, LOU Hu, YANG Guang, SUN Long. Changes of fine root functional traits and rhizosphere bacterial community of Betula platyphylla after fire [J]. Chin J Plant Ecol, 2024, 48(7): 828-843. |
| [3] | JIAO Hui-Ying, LIU Li-Qiang, YANG Jia-Xin, QIN Wei, WANG Rui-Zhe. Effects of rhizosphere nitrogen-fixing, phosphate-solubilizing and potassium-solubilizing bacteria on leaf nutrients and physiological traits in different natural populations of Malus sieversii [J]. Chin J Plant Ecol, 2024, 48(7): 930-942. |
| [4] | PENG Zhong-Tao, JIN Guang-Ze, LIU Zhi-Li. Leaf trait variations and relationships of three Acer species in different tree sizes and canopy conditions in Xiao Hinggan Mountains of Northeast China [J]. Chin J Plant Ecol, 2024, 48(6): 730-743. |
| [5] | FU Liang-Chen, DING Zong-Ju, TANG Mao, ZENG Hui, ZHU Biao. Rhizosphere effects of Betula platyphylla and Quercus mongolica and their seasonal dynamics in Dongling Mountain, Beijing [J]. Chin J Plant Ecol, 2024, 48(4): 508-522. |
| [6] | FAN Hong-Kun, ZENG Tao, JIN Guang-Ze, LIU Zhi-Li. Leaf trait variation and trade-offs among growth types of broadleaf plants in Xiao Hinggan Mountains [J]. Chin J Plant Ecol, 2024, 48(3): 364-376. |
| [7] | HUANG Ling, WANG Zhen, MA Ze, YANG Fa-Lin, LI Lan, SEREKPAYEV Nurlan, NOGAYEV Adilbek, HOU Fu-Jiang. Effects of long-term grazing and nitrogen addition on the growth of Stipa bungeana population in typical steppe of Loess Plateau [J]. Chin J Plant Ecol, 2024, 48(3): 317-330. |
| [8] | Ma Bin Wei-Wei SHE Qin Huan Song Chunyang Yuan Xinyue Chun MIAO Liu Liang Feng Wei Qin shugao Yuqing Zhang. Effects of nitrogen and water addition on seed functional traits of Artemisia ordosica [J]. Chin J Plant Ecol, 2024, 48(12): 1637-1649. |
| [9] | HU Chu-Ting, YANG Liu-Yi-Yi, SHI Shao-Lin, ZHOU Yan, CHEN Ting-Ting, ZHENG Bo-Han, YANG Yang, LU Xiao-Ling, WANG Chen-Ling, Ni Jian. Plant functional traits of typical artificial vegetation in Jinhua, Zhejiang, China [J]. Chin J Plant Ecol, 2024, 48(10): 1336-1350. |
| [10] | Zumureti YUSUFUJANG, DONG Zheng-Wu, CHENG Peng, YE Mao, LIU Sui-Yun-Hao, LI Sheng-Yu, ZHAO Xiao-Ying. Response of water use strategies of Tamarix ramosissima to nebkhas accumulation process [J]. Chin J Plant Ecol, 2024, 48(1): 113-126. |
| [11] | ZHANG Zhi-Shan, HAN Gao-Ling, HUO Jian-Qiang, HUANG Ri-Hui, XUE Shu-Wen. Response of xylem hydraulic conductivity and leaf photosynthetic capacity of sand-binding shrubs Caragana korshinskii and C. liouana to soil water [J]. Chin J Plant Ecol, 2023, 47(10): 1422-1431. |
| [12] | DONG Liu-Wen, REN Zheng-Wei, ZHANG Rui, XIE Chen-Di, ZHOU Xiao-Long. Functional diversity rather than species diversity can explain community biomass variation following short-term nitrogen addition in an alpine grassland [J]. Chin J Plant Ecol, 2022, 46(8): 871-881. |
| [13] | ZANG Yong-Xin, MA Jian-Ying, ZHOU Xiao-Bing, TAO Ye, YIN Ben-Feng, Shayaguli JIGEER, ZHANG Yuan-Ming. Effects of extreme drought and extreme precipitation on aboveground productivity of ephemeral plants across different slope positions along sand dunes [J]. Chin J Plant Ecol, 2022, 46(12): 1537-1550. |
| [14] | Min FAN, Yi-Tong LU, Zhao-Hua WANG, Ying-Qi HUANG, Yu PENG, Jia-Xin SHANG, Yang ZHANG. Effects of patch pattern on plant diversity and functional traits in center Hunshandak Sandland [J]. Chin J Plant Ecol, 2022, 46(1): 51-61. |
| [15] | WANG Chun-Cheng, ZHANG Yun-Ling, MA Song-Mei, HUANG Gang, ZHANG Dan, YAN Han. Phylogeny and species differentiation of four wild almond species of subgen. Amygdalus in China [J]. Chin J Plant Ecol, 2021, 45(9): 987-995. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Chinese Journal of Plant Ecology
Tel: 010-62836134, 62836138, E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn