

Chin J Plan Ecolo ›› 2015, Vol. 39 ›› Issue (4): 383-387.DOI: 10.17521/cjpe.2015.0037
• Orginal Article • Previous Articles Next Articles
YANG Hai-Shui*( ), WANG Qi, GUO Yi, XIONG Yan-Qin, XU Ming-Min, DAI Ya-Jun
), WANG Qi, GUO Yi, XIONG Yan-Qin, XU Ming-Min, DAI Ya-Jun
Received:2014-10-11
Accepted:2015-01-17
Online:2015-04-01
Published:2015-04-21
Contact:
Hai-Shui YANG
About author:# Co-first authors
YANG Hai-Shui,WANG Qi,GUO Yi,XIONG Yan-Qin,XU Ming-Min,DAI Ya-Jun. Correlation analysis between arbuscular mycorrhizal fungal community and host plant phylogeny[J]. Chin J Plan Ecolo, 2015, 39(4): 383-387.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.plant-ecology.com/EN/10.17521/cjpe.2015.0037
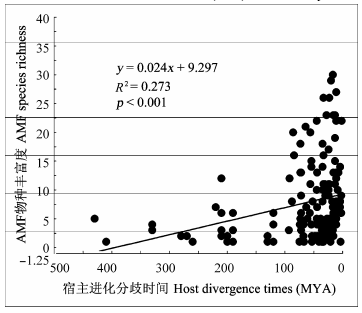
Fig. 1 Relationship between the time of divergence of hosts (MYA, million year) and the richness of arbuscular mycorrhizal fungal (AMF) species in roots.
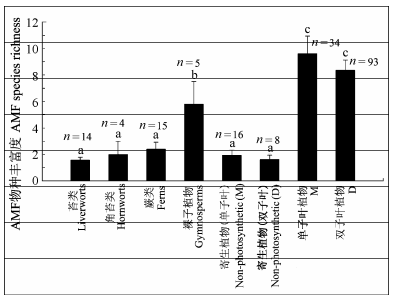
Fig. 2 Distribution of species richness of arbuscular mycorrhizal fungi (AMF) in different plant phylogenetic groups (mean ± SE). Different lowercase letters indicate significant differences among different plant phylogenetic groups (p < 0.05). D, dicotyledons; M, monocotyledons.
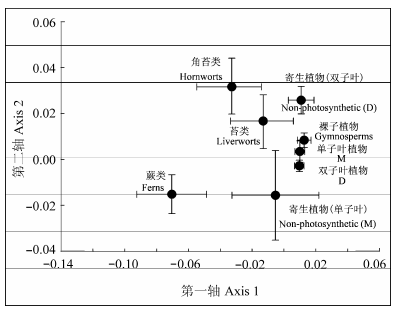
Fig. 3 Non-metric multidimensional scaling (NMDS) analysis of arbuscular mycorrhizal fungal community composition in different phylogenetic groups. D, dicotyledons; M, monocotyledons.
| L | H | F | G | NPM | M | NPD | D | |
|---|---|---|---|---|---|---|---|---|
| L | - | 0.02 | <0.01 | 0.46 | <0.01 | <0.01 | <0.01 | <0.01 |
| H | - | 0.02 | 0.02 | 0.01 | <0.01 | <0.01 | <0.01 | |
| F | - | 0.07 | 0.01 | <0.01 | <0.01 | <0.01 | ||
| G | - | 0.21 | 0.58 | <0.01 | 0.68 | |||
| NPM | - | <0.01 | <0.01 | <0.01 | ||||
| M | - | <0.01 | 0.94 | |||||
| NPD | - | <0.01 | ||||||
| D | - |
Table 1 One-way NPMANOVA of arbuscular mycorrhizal fungal community composition in different phylogenetic groups of host plants
| L | H | F | G | NPM | M | NPD | D | |
|---|---|---|---|---|---|---|---|---|
| L | - | 0.02 | <0.01 | 0.46 | <0.01 | <0.01 | <0.01 | <0.01 |
| H | - | 0.02 | 0.02 | 0.01 | <0.01 | <0.01 | <0.01 | |
| F | - | 0.07 | 0.01 | <0.01 | <0.01 | <0.01 | ||
| G | - | 0.21 | 0.58 | <0.01 | 0.68 | |||
| NPM | - | <0.01 | <0.01 | <0.01 | ||||
| M | - | <0.01 | 0.94 | |||||
| NPD | - | <0.01 | ||||||
| D | - |
| [1] | Bidartondo MI, Redecker D, Hijri I, Wiemken A, Bruns TD, Domínguez L, Sersic A, Leake JR, Read DJ (2002). Epiparasitic plants specialized on arbuscular mycorrhizal fungi.Nature, 419, 389-392. |
| [2] | Borowicz VA (2001). Do arbuscular mycorrhizal fungi alter plant-pathogen relations? Ecology, 82, 3057-3068. |
| [3] | Field KJ, Cameron DD, Leake JR, Tille S, Bidartondo MI, Beerling DJ (2012). Contrasting arbuscular mycorrhizal responses of vascular and non-vascular plants to a simulated palaeozoic CO2 decline.Nature Communica- tions, 3, 1-8. |
| [4] | Fiz-Palacios O, Schneider H, Heinrichs J, Savolainen V (2011). Diversification of land plants: Insights from a family-level phylogenetic analysis.BMC Evolutionary Biology, 11, 341. |
| [5] | Fonseca HMAC, Berbara RLL (2008). Does Lunularia cruciata form symbiotic relationships with either Glomus proliferum or G. intraradices?Mycological Research, 112, 1063-1068. |
| [6] | Franke T, Beenken L, Döring M, Kocyan A, Agerer R (2006). Arbuscular mycorrhizal fungi of the Glomus-group A lineage (Glomerales; Glomeromycota) detected in myco- heterotrophic plants from tropical Africa.Mycological Progress, 5, 24-31. |
| [7] | Hammer Ø, Harper DAT, Ryan PD (2001). PAST: Paleontolo- gical statistics software package for education and data analysis.Palaeontologia Electronica, 4, 1-9. |
| [8] | Humphreys CP, Franks PJ, Rees M, Bidartondo MI, Leake JR, Beerling DJ (2010). Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants.Nature Communications, 1, 103. |
| [9] | Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E, Fellbaum CR, Kowalchuk GA, Hart MM, Bago A (2011). Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis.Science, 333, 880-882. |
| [10] | Koide R, Dickie I (2002). Effects of mycorrhizal fungi on plant populations.Plant and Soil, 244, 307-317. |
| [11] | Kottke I, Nebel M (2005). The evolution of mycorrhiza-like associations in liverworts: An update.New Phytologist, 167, 330-334. |
| [12] | Kovács GM, Balázs T, Pénzes Z (2007). Molecular study of arbuscular mycorrhizal fungi colonizing the sporophyte of the eusporangiate rattlesnake fern (Botrychium virginian- um, Ophioglossaceae). Mycorrhiza, 17, 597-605. |
| [13] | Ligrone R, Carafa A, Lumini E, Bianciotto V, Bonfante P, Duckett JG (2007). Glomeromycotean associations in liverworts: A molecular, cellular, and taxonomic analysis.American Journal of Botany, 94, 1756-1777. |
| [14] | Öpik M, Vanatoa A, Vanatoa E, Moora M, Davison J, Kalwij JM, Reier Ü, Zobel M (2010). The online database Maarj AM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromy- cota).New Phytologist, 188, 223-241. |
| [15] | Pirozynski KA, Malloch DW (1975). The origin of land plants: Amatter of mycotrophism.Biosystems, 6, 153-164. |
| [16] | Read D, Duckett J, Francis R, Ligrone R, Russell A (2000). Symbiotic fungal associations in “lower” land plants.Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences, 355, 815-831. |
| [17] | Redecker D, Kodner R, Graham LE (2000). Glomalean fungi from the Ordovician.Science, 289, 1920-1921. |
| [18] | Remy W, Taylor TN, Hass H, Kerp H (1994). Four hundred- million-year-old vesicular arbuscular mycorrhizae.Proceedings of the National Academy of Sciences of the United of America, 91, 11841-11843. |
| [19] | Russell J, Bulman S (2005). The liverwort Marchantia foliacea forms a specialized symbiosis with arbuscular mycorrhizal fungi in the genus Glomus.New Phytologist, 165, 567-579. |
| [20] | Sanderson MJ (2003). r8s: Inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock.Bioinformatics, 19, 301-302. |
| [21] | Smith SE, Read DJ (2008). Mycorrhizal Symbiosis. 3rd edn. Academic Press, San Diego, USA. |
| [22] | Stamatakis A (2006). RAxML-VI-HPC: Maximum likelihood- based phylogenetic analyses with thousands of taxa and mixed models.Bioinformatics, 22, 2688-2690. |
| [23] | van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, Wiemken A, Sanders IR (1998). Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity.Nature, 396, 69-72. |
| [24] | van der Heijden MGA, Scheublin TR, Brader A (2004). Taxon- omic and functional diversity in arbuscular mycorrhizal fungi―Is there any relationship?New Phytologist, 164, 201-204. |
| [25] | Wagg C, Jansa J, Stadler M, Schmid B, van der Heijden MGA (2011). Mycorrhizal fungal identity and diversity relaxes plant-plant competition.Ecology, 92, 1303-1313. |
| [26] | Wang B, Yeun LH, Xue JY, Liu Y, Ané JM, Qiu YL (2010). Presence of three mycorrhizal genes in the common ancestor of land plants suggests a key role of mycorrhizas in the colonization of land by plants.New Phytologist, 186, 514-525. |
| [27] | Winther JL, Friedman WE (2007). Arbuscular mycorrhizal symbionts in Botrychium (Ophioglossaceae).American Journal of Botany, 94, 1248-1255. |
| [28] | Wu JP, Liu ZF, Wang XL, Sun YX, Zhou LX, Lin YB, Fu SL (2011). Effects of understory removal and tree girdling on soil microbial community composition and litter decomposition in two Eucalyptus plantations in South China.Functional Ecology, 25, 921-931. |
| [1] | Ke-Yu CHEN Sen Xing Yu Tang Sun JiaHui Shijie Ren Bao-Ming JI. Arbuscular mycorrhizal fungal community characteristics and driving factors in different grassland types [J]. Chin J Plant Ecol, 2024, 48(5): 660-674. |
| [2] | Die Hu Xinqi Jiang DAI Zhicong Daiyi Chen Yu Zhang Shan-Shan Qi. Arbuscular mycorrhizal fungi enhance the herbicide tolerance of an invasive weed Sphagneticola trilobata [J]. Chin J Plant Ecol, 2024, 48(5): 651-659. |
| [3] | YANG An-Na, LI Zeng-Yan, MOU Ling, YANG Bai-Yu, SAI Bi-Le, ZHANG Li, ZHANG Zeng-Ke, WANG Wan-Sheng, DU Yun-Cai, YOU Wen-Hui, YAN En-Rong. Variation in soil bacterial community across vegetation types in Dajinshan Island, Shanghai [J]. Chin J Plant Ecol, 2024, 48(3): 377-389. |
| [4] | NIU Yi-Di, CAI Ti-Jiu. Changes in species diversity and influencing factors in secondary forest succession in northern Da Hinggan Mountains [J]. Chin J Plant Ecol, 2024, 48(3): 349-363. |
| [5] | CHEN Zhao-Quan, WANG Ming-Hui, HU Zi-Han, LANG Xue-Dong, HE Yun-Qiong, LIU Wan-De. Mechanisms of seedling community assembly in a monsoon evergreen broadleaf forest in Pu’er, Yunnan, China [J]. Chin J Plant Ecol, 2024, 48(1): 68-79. |
| [6] | CHEN Bao-Dong, FU Wei, WU Song-Lin, ZHU Yong-Guan. Involvements of mycorrhizal fungi in terrestrial ecosystem carbon cycling [J]. Chin J Plant Ecol, 2024, 48(1): 1-20. |
| [7] | YANG Ming-Wei, JIN Xiao-Fang. Diversity and evolutionary ecology of nectar spurs in angiosperms [J]. Chin J Plant Ecol, 2023, 47(9): 1193-1210. |
| [8] | LI Na, TANG Shi-Ming, GUO Jian-Ying, TIAN Ru, WANG Shan, HU Bing, LUO Yong-Hong, XU Zhu-Wen. Meta-analysis of effects of grazing on plant community properties in Nei Mongol grassland [J]. Chin J Plant Ecol, 2023, 47(9): 1256-1269. |
| [9] | YANG Xin, REN Ming-Xun. Species distribution pattern and formation mechanism of mangrove plants around the South China Sea [J]. Chin J Plant Ecol, 2023, 47(8): 1105-1115. |
| [10] | YU Xiao, JI Ruo-Xuan, REN Tian-Meng, XIA Xin-Li, YIN Wei-Lun, LIU Chao. Distribution, characteristics and classification of Caryopteris mongholica communities in northern China [J]. Chin J Plant Ecol, 2023, 47(8): 1182-1192. |
| [11] | ZHANG Zhong-Yang, SONG Xi-Qiang, REN Ming-Xun, ZHANG Zhe. Ecological functions of vascular epiphytes in habitat construction [J]. Chin J Plant Ecol, 2023, 47(7): 895-911. |
| [12] | HE Fei, LI Chuan, Faisal SHAH, LU Xie-Min, WANG Ying, WANG Meng, RUAN Jia, WEI Meng-Lin, MA Xing-Guang, WANG Zhuo, JIANG Hao. Carbon transport and phosphorus uptake in an intercropping system of Robinia pseudoacacia and Amorphophallus konjac mediated by arbuscular mycorrhizal hyphal networks [J]. Chin J Plant Ecol, 2023, 47(6): 782-791. |
| [13] | YANG Jia-Rong, DAI Dong, CHEN Jun-Fang, WU Xian, LIU Xiao-Lin, LIU Yu. Insight into recent studies on the diversity of arbuscular mycorrhizal fungi in shaping plant community assembly and maintaining rare species [J]. Chin J Plant Ecol, 2023, 47(6): 745-755. |
| [14] | HU Tong-Xin, LI Bei, LI Guang-Xin, REN Yue-Xiao, DING Hai-Lei, SUN Long. Effects of fire originated black carbon on species composition of ectomycorrhizal fungi in a Larix gmelinii forest in growing season [J]. Chin J Plant Ecol, 2023, 47(6): 792-803. |
| [15] | LÜ Zi-Li, LIU Bin, CHANG Feng, MA Zi-Jing, CAO Qiu-Mei. Relationship between plant functional diversity and ecosystem multifunctionality in Bayanbulak alpine meadow along an altitude gradient [J]. Chin J Plant Ecol, 2023, 47(6): 822-832. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Chinese Journal of Plant Ecology
Tel: 010-62836134, 62836138, E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn