

植物生态学报 ›› 2022, Vol. 46 ›› Issue (3): 249-266.DOI: 10.17521/cjpe.2021.0059
• 综述 • 下一篇
谢育杭1,*, 贾璞1,*, 郑修坛1, 李金天1, 束文圣1, 王宇涛1,2,**( )
)
收稿日期:2021-02-22
接受日期:2021-08-02
出版日期:2022-03-20
发布日期:2021-08-27
通讯作者:
王宇涛
作者简介:** (wangyutao@scnu.edu.cn)* 同等贡献
基金资助:
XIE Yu-Hang1,*, JIA Pu1,*, ZHENG Xiu-Tan1, LI Jin-Tian1, SHU Wen-Sheng1, WANG Yu-Tao1,2,**( )
)
Received:2021-02-22
Accepted:2021-08-02
Online:2022-03-20
Published:2021-08-27
Contact:
WANG Yu-Tao
About author:First author contact:* Contributed equally to this work
Supported by:摘要:
植物与共存微生物的相互作用对植物的生长、发育、健康等具有重大影响。人类驯化导致现代作物品种与其野生祖先在生理遗传特性、生长环境等方面存在明显差异, 这必然会影响作物与其微生物组的相互作用。理解驯化对作物微生物组的影响及其作用机理, 是充分应用微生物组进行作物改良或人工育种的重要理论基础。结合课题组前期研究基础, 该文综述了驯化对作物地下和地上部分细菌和真菌(尤其是益生菌和病原菌)群落组成和多样性影响的研究现状; 并结合驯化对作物植株形态、根系构型、根系分泌物等生理特征以及生长环境的影响, 分析了驯化塑造作物微生物组的作用途径, 提出了该领域值得重点关注的研究和发展方向。
谢育杭, 贾璞, 郑修坛, 李金天, 束文圣, 王宇涛. 驯化对作物微生物组多样性和群落结构的影响及作用途径. 植物生态学报, 2022, 46(3): 249-266. DOI: 10.17521/cjpe.2021.0059
XIE Yu-Hang, JIA Pu, ZHENG Xiu-Tan, LI Jin-Tian, SHU Wen-Sheng, WANG Yu-Tao. Impacts and action pathways of domestication on diversity and community structure of crop microbiome: a review. Chinese Journal of Plant Ecology, 2022, 46(3): 249-266. DOI: 10.17521/cjpe.2021.0059
| 作物 Crop | 部位 Tissue | 细菌 Bacteria | 真菌 Fungi | 参考文献 Reference | ||
|---|---|---|---|---|---|---|
| 多样性 Diversity | 群落结构 Community structure | 多样性 Diversity | 群落结构 Community structure | |||
| 玉米 Zea mays | 根 Root | ↓ | S. | ↓ | S. | Sangabriel-Conde et al., |
| 种子 Seed | N.S. | S. | - | - | Johnston-Monje & Raizada, | |
| 水稻 Oryza sativa | 根 Root | ↓ | S. | ? | S. | Shenton et al., Shi et al., |
| 种子 Seed | - | S. | - | S. | Kim et al., | |
| 小麦 Triticum aestivum | 根 Root | ↓ | S. | ↓ | S. | Ofek-Lalzar et al., |
| 种子 Seed | N.S. | S. | N.S. | S. | Özkurt et al., | |
| 叶 Leaf | ↓ | S. | N.S. | S. | Hassani et al., | |
| 大豆 Glycine max | 根 Root | ↓ | S. | ? | S. | Liu et al., |
| 甜菜 Beta vulgaris | 根 Root | ↓ | S. | - | - | Zachow et al., |
| 向日葵 Helianthus annuus | 根 Root | N.S. | N.S. | N.S. | S. | Leff et al., |
| 小米 Setaria italica | 根 Root | N.S. | S. | - | - | Chaluvadi & Bennetzen, |
| 菜豆 Phaseolus vulgaris | 根 Root | ↑ | S. | - | - | Pérez-Jaramillo et al., |
| Agave tequilana | 根 Root | ↓ | S. | ? | S. | Coleman-Derr et al., |
| 叶 Leaf | ↓ | S. | ? | S. | ||
| 面包树 Artocarpus communis | 根 Root | - | - | ↓ | S. | Xing et al., |
表1 驯化(野生种vs.栽培种)对不同作物细菌、真菌多样性和群落结构的影响
Table 1 Effects of domestication (wild vs. cultivated) on the diversity and community structure of bacteria and fungi in different crops
| 作物 Crop | 部位 Tissue | 细菌 Bacteria | 真菌 Fungi | 参考文献 Reference | ||
|---|---|---|---|---|---|---|
| 多样性 Diversity | 群落结构 Community structure | 多样性 Diversity | 群落结构 Community structure | |||
| 玉米 Zea mays | 根 Root | ↓ | S. | ↓ | S. | Sangabriel-Conde et al., |
| 种子 Seed | N.S. | S. | - | - | Johnston-Monje & Raizada, | |
| 水稻 Oryza sativa | 根 Root | ↓ | S. | ? | S. | Shenton et al., Shi et al., |
| 种子 Seed | - | S. | - | S. | Kim et al., | |
| 小麦 Triticum aestivum | 根 Root | ↓ | S. | ↓ | S. | Ofek-Lalzar et al., |
| 种子 Seed | N.S. | S. | N.S. | S. | Özkurt et al., | |
| 叶 Leaf | ↓ | S. | N.S. | S. | Hassani et al., | |
| 大豆 Glycine max | 根 Root | ↓ | S. | ? | S. | Liu et al., |
| 甜菜 Beta vulgaris | 根 Root | ↓ | S. | - | - | Zachow et al., |
| 向日葵 Helianthus annuus | 根 Root | N.S. | N.S. | N.S. | S. | Leff et al., |
| 小米 Setaria italica | 根 Root | N.S. | S. | - | - | Chaluvadi & Bennetzen, |
| 菜豆 Phaseolus vulgaris | 根 Root | ↑ | S. | - | - | Pérez-Jaramillo et al., |
| Agave tequilana | 根 Root | ↓ | S. | ? | S. | Coleman-Derr et al., |
| 叶 Leaf | ↓ | S. | ? | S. | ||
| 面包树 Artocarpus communis | 根 Root | - | - | ↓ | S. | Xing et al., |
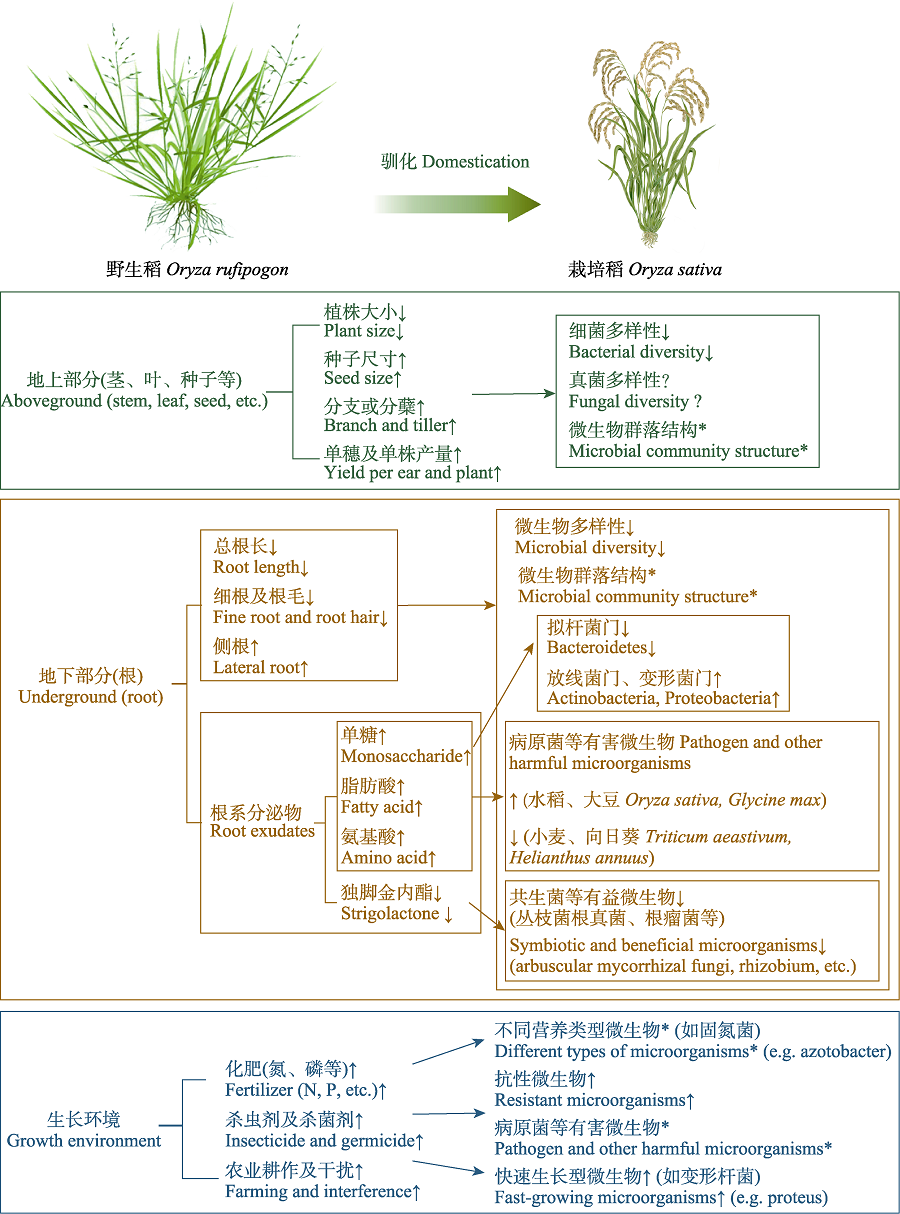
图1 驯化对作物(以水稻为例)微生物组群落组成和多样性的作用途径。↑、↓分别表示显著增加和降低; *表示有显著差异; ?表示尚不明确。
Fig. 1 Action pathways of domestication on the composition and diversity of microbiome in crops (e.g. rice). “↑” and “↓” indicate significant increase and decrease, respectively; “*” indicates significant difference; “?” indicates not yet clear. N, nitrogen; P, phosphorus.
| [1] |
Abbo S, Gopher A (2017). Near eastern plant domestication: a history of thought. Trends in Plant Science, 22, 491-511.
DOI URL |
| [2] |
Abbo S, Pinhasi van-Oss R, Gopher A, Saranga Y, Ofner I, Peleg Z (2014). Plant domestication versus crop evolution: a conceptual framework for cereals and grain legumes. Trends in Plant Science, 19, 351-360.
DOI URL |
| [3] |
Abboud MAA (2014). Bioimpact of application of pesticides with plant growth hormone (gibberellic acid) on target and non-target microorganisms. Journal of Saudi Chemical Society, 18, 1005-1010.
DOI URL |
| [4] |
Alegria Terrazas R, Balbirnie-Cumming K, Morris J, Hedley PE, Russell J, Paterson E, Baggs EM, Fridman E, Bulgarelli D (2020). A footprint of plant eco-geographic adaptation on the composition of the barley rhizosphere bacterial microbiota. Scientific Reports, 10, 12916. DOI: 10.1038/s41598-020-69672-x.
DOI PMID |
| [5] |
Armalytė J, Skerniškytė J, Bakienė E, Krasauskas R, Šiugždinienė R, Kareivienė V, Kerzienė S, Klimienė I, Sužiedėlienė E, Ružauskas M (2019). Microbial diversity and antimicrobial resistance profile in microbiota from soils of conventional and organic farming systems. Frontiers in Microbiology, 10, 892. DOI: 10.3389/fmicb.2019.00892.
DOI PMID |
| [6] | Badri DV, Vivanco JM (2009). Regulation and function of root exudates. Plant, Cell & Environment, 32, 666-681. |
| [7] | Banik A, Kumar U, Mukhopadhyay SK, Dangar TK (2017). Dynamics of endophytic and epiphytic bacterial communities of Indian cultivated and wild rice (Oryza spp.) genotypes. Ecological Genetics and Genomics, 3-5, 7-17. |
| [8] |
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012). The rhizosphere microbiome and plant health. Trends in Plant Science, 17, 478-486.
DOI PMID |
| [9] |
Berg G, Köberl M, Rybakova D, Müller H, Grosch R, Smalla K (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiology Ecology, 93, fix050. DOI: 10.1093/femsec/fix050.
DOI |
| [10] |
Berg G, Raaijmakers JM (2018). Saving seed microbiomes. The ISME Journal, 12, 1167-1170.
DOI URL |
| [11] |
Berg G, Rybakova D, Grube M, Köberl M (2016). The plant microbiome explored: implications for experimental botany. Journal of Experimental Botany, 67, 995-1002.
DOI URL |
| [12] |
Bodenhausen N, Bortfeld-Miller M, Ackermann M, Vorholt JA (2014). A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLOS Genetics, 10, e1004283. DOI: 10.1371/journal.pgen.1004283.
DOI URL |
| [13] |
Bourion V, Heulin-Gotty K, Aubert V, Tisseyre P, Chabert-Martinello M, Pervent M, Delaitre C, Vile D, Siol M, Duc G, Brunel B, Burstin J, Lepetit M (2018). Co-inoculation of a pea core-collection with diverse rhizobial strains shows competitiveness for nodulation and efficiency of nitrogen fixation are distinct traits in the interaction. Frontiers in Plant Science, 8, 2249. DOI: 10.3389/fpls.2017.02249.
DOI URL |
| [14] |
Brader G, Compant S, Mitter B, Trognitz F, Sessitsch A (2014). Metabolic potential of endophytic bacteria. Current Opinion in Biotechnology, 27, 30-37.
DOI URL |
| [15] |
Brisson VL, Schmidt JE, Northen TR, Vogel JP, Gaudin ACM (2019). Impacts of maize domestication and breeding on rhizosphere microbial community recruitment from a nutrient depleted agricultural soil. Scientific Reports, 9, 15611. DOI: 10.1038/s41598-019-52148-y.
DOI URL |
| [16] |
Busby PE, Soman C, Wagner MR, Friesen ML, Kremer J, Bennett A, Morsy M, Eisen JA, Leach JE, Dangl JL (2017). Research priorities for harnessing plant microbiomes in sustainable agriculture. PLOS Biology, 15, e2001793. DOI: 10.1371/journal.pbio.2001793.
DOI URL |
| [17] |
Carrión VJ, Perez-Jaramillo J, Cordovez V, Tracanna V, de Hollander M, Ruiz-Buck D, Mendes LW, Gomez-Exposito R, Elsayed SS, Mohanraju P, Arifah A, van der Oost J, Paulson JN, Mendes R, et al. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science, 366, 606-612.
DOI PMID |
| [18] |
Cassman NA, Leite MF, Pan Y, de Hollander M, van Veen JA, Kuramae EE (2016). Plant and soil fungal but not soil bacterial communities are linked in long-term fertilized grassland. Scientific Reports, 6, 23680. DOI: 10.1038/srep23680.
DOI PMID |
| [19] |
Chaluvadi S, Bennetzen JL (2018). Species-associated differences in the below-ground microbiomes of wild and domesticated Setaria. Frontiers in Plant Science, 9, 1183. DOI: 10.3389/fpls.2018.01183.
DOI URL |
| [20] |
Chang CL, Chen W, Luo SS, Ma LN, Li XJ, Tian CJ (2019). Rhizosphere microbiota assemblage associated with wild and cultivated soybeans grown in three types of soil suspensions. Archives of Agronomy and Soil Science, 65, 74-87.
DOI URL |
| [21] |
Chang CL, Zhang JX, Liu TT, Song KJ, Xie JH, Luo SS, Qu TB, Zhang JJ, Tian CJ, Zhang JF (2020). Rhizosphere fungal communities of wild and cultivated soybeans grown in three different soil suspensions. Applied Soil Ecology, 153, 103586. DOI: 10.1016/j.apsoil.2020.103586.
DOI URL |
| [22] |
Chen L, Brookes PC, Xu JM, Zhang JB, Zhang CZ, Zhou XY, Luo Y (2016). Structural and functional differentiation of the root-associated bacterial microbiomes of perennial ryegrass. Soil Biology & Biochemistry, 98, 1-10.
DOI URL |
| [23] |
Chen LX, Méndez-García C, Dombrowski N, Servín-Garcidueñas LE, Eloe-Fadrosh EA, Fang BZ, Luo ZH, Tan S, Zhi XY, Hua ZS, Martinez-Romero E, Woyke T, Huang LN, Sánchez J, Peláez AI, et al. (2018a). Metabolic versatility of small archaea Micrarchaeota and Parvarchaeota. The ISME Journal, 12, 756-775.
DOI URL |
| [24] |
Chen YH, Bernal CC (2011). Arthropod diversity and community composition on wild and cultivated rice. Agricultural and Forest Entomology, 13, 181-189.
DOI URL |
| [25] |
Chen YH, Gols R, Benrey B (2015). Crop domestication and its impact on naturally selected trophic interactions. Annual Review of Entomology, 60, 35-58.
DOI URL |
| [26] |
Chen YH, Ruiz-Arocho J, von Wettberg EJ (2018b). Crop domestication: anthropogenic effects on insect-plant interactions in agroecosystems. Current Opinion in Insect Science, 29, 56-63.
DOI URL |
| [27] |
Coleman-Derr D, Desgarennes D, Fonseca-Garcia C, Gross S, Clingenpeel S, Woyke T, North G, Visel A, Partida-Martinez LP, Tringe SG (2016). Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytologist, 209, 798-811.
DOI PMID |
| [28] |
Compant S, Samad A, Faist H, Sessitsch A (2019). A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. Journal of Advanced Research, 19, 29-37.
DOI URL |
| [29] |
Cordovez V, Dini-Andreote F, Carrión VJ, Raaijmakers JM (2019). Ecology and evolution of plant microbiomes. Annual Review of Microbiology, 73, 69-88.
DOI PMID |
| [30] |
Corneo PE, Suenaga H, Kertesz MA, Dijkstra FA (2016). Effect of twenty four wheat genotypes on soil biochemical and microbial properties. Plant and Soil, 404, 141-155.
DOI URL |
| [31] |
da Fonseca RR, Smith BD, Wales N, Cappellini E, Skoglund P, Fumagalli M, Samaniego JA, Carøe C, Ávila-Arcos MC, Hufnagel DE, Korneliussen TS, Vieira FG, Jakobsson M, Arriaza B, Willerslev E, et al. (2015). The origin and evolution of maize in the Southwestern United States. Nature Plants, 1, 14003. DOI: 10.1038/nplants.2014.3.
DOI PMID |
| [32] |
da Silva K, da Silva EE, Farias ENC, Chaves JS, Albuquerque CNB, Cardoso C (2018). Agronomic efficiency of Bradyrhizobium pre-inoculation in association with chemical treatment of soybean seeds. African Journal of Agricultural Research, 13, 726-732.
DOI URL |
| [33] |
de Vries FT, Griffiths RI, Knight CG, Nicolitch O, Williams A (2020). Harnessing rhizosphere microbiomes for drought- resilient crop production. Science, 368, 270-274.
DOI URL |
| [34] | Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proceedings of the National Academy of Sciences of the United States of America, 112, E911-E920. |
| [35] |
Emmett BD, Buckley DH, Smith ME, Drinkwater LE (2018). Eighty years of maize breeding alters plant nitrogen acquisition but not rhizosphere bacterial community composition. Plant and Soil, 431, 53-69.
DOI URL |
| [36] |
Fan K, Delgado-Baquerizo M, Guo X, Wang D, Wu Y, Zhu M, Yu W, Yao H, Zhu YG, Chu H (2019). Suppressed N fixation and diazotrophs after four decades of fertilization. Microbiome, 7, 143. DOI: 10.1186/s40168-019-0757-8.
DOI URL |
| [37] |
Faris JD, Zhang Q, Chao S, Zhang Z, Xu SS (2014). Analysis of agronomic and domestication traits in a durum × cultivated emmer wheat population using a high-density single nucleotide polymorphism-based linkage map. Theoretical and Applied Genetics, 127, 2333-2348.
DOI URL |
| [38] |
Firrincieli A, Khorasani M, Frank AC, Doty SL (2020). Influences of climate on phyllosphere endophytic bacterial communities of wild poplar. Frontiers in Plant Science, 11, 203. DOI: 10.3389/fpls.2020.00203.
DOI PMID |
| [39] |
Francioli D, Schulz E, Lentendu G, Wubet T, Buscot F, Reitz T (2016). Mineral vs. organic amendments: microbial community structure, activity and abundance of agriculturally relevant microbes are driven by long-term fertilization strategies. Frontiers in Microbiology, 7, 1446. DOI: 10.3389/fmicb.2016.01446.
DOI PMID |
| [40] |
Haas M, Schreiber M, Mascher M (2019). Domestication and crop evolution of wheat and barley: genes, genomics, and future directions. Journal of Integrative Plant Biology, 61, 204-225.
DOI URL |
| [41] |
Hassan S, Mathesius U (2012). The role of flavonoids in root-rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. Journal of Experimental Botany, 63, 3429-3444.
DOI URL |
| [42] |
Hassani MA, Özkurt E, Franzenburg S, Stukenbrock EH (2020). Ecological assembly processes of the bacterial and fungal microbiota of wild and domesticated wheat species. Phytobiomes Journal, 4, 217-224.
DOI URL |
| [43] |
Hassani MA, Özkurt E, Seybold H, Dagan T, Stukenbrock EH (2019). Interactions and coadaptation in plant metaorganisms. Annual Review of Phytopathology, 57, 483-503.
DOI PMID |
| [44] |
Horton MW, Bodenhausen N, Beilsmith K, Meng D, Muegge BD, Subramanian S, Vetter MM, Vilhjálmsson BJ, Nordborg M, Gordon JI, Bergelson J (2014). Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nature Communications, 5, 5320. DOI: 10.1038/ncomms6320.
DOI URL |
| [45] |
Hu M, Lv SW, Wu WG, Fu YC, Liu FX, Wang BB, Li WG, Gu P, Cai HW, Sun CQ, Zhu ZF (2018). The domestication of plant architecture in African rice. The Plant Journal, 94, 661-669.
DOI URL |
| [46] |
Huang LN, Kuang JL, Shu WS (2016). Microbial ecology and evolution in the acid mine drainage model system. Trends in Microbiology, 24, 581-593.
DOI URL |
| [47] |
Hunter PJ, Pink DAC, Bending GD (2015). Cultivar-level genotype differences influence diversity and composition of lettuce (Lactuca sp.) phyllosphere fungal communities. Fungal Ecology, 17, 183-186.
DOI URL |
| [48] |
Iannucci A, Fragasso M, Beleggia R, Nigro F, Papa R (2017). Evolution of the crop rhizosphere: impact of domestication on root exudates in tetraploid wheat (Triticum turgidum L.). Frontiers in Plant Science, 8, 2124. DOI: 10.3389/fpls.2017.02124.
DOI PMID |
| [49] |
Ikeda-Ohtsubo W, Brugman S, Warden CH, Rebel JMJ, Folkerts G, Pieterse CMJ (2018). How can we define “optimal microbiota?”: a comparative review of structure and functions of microbiota of animals, fish, and plants in agriculture. Frontiers in Nutrition, 5, 90. DOI: 10.3389/fnut.2018.00090.
DOI PMID |
| [50] | Ji L, Shi SH, Tian L, Tian CJ (2019). A discussion on issues related to the recruit of rhizosphere microbes caused by crop domestication. Soils and Crops, 8, 368-372. |
| [吉丽, 石少华, 田磊, 田春杰 (2019). 作物驯化对根际微生物组的选择. 土壤与作物, 8, 368-372.] | |
| [51] |
Jiang Y, Luan L, Hu K, Liu M, Chen Z, Geisen S, Chen X, Li H, Xu Q, Bonkowski M, Sun B (2020). Trophic interactions as determinants of the arbuscular mycorrhizal fungal community with cascading plant-promoting consequences. Microbiome, 8, 142. DOI: 10.1186/s40168-020-00918-6.
DOI PMID |
| [52] |
Johnston-Monje D, Mousa WK, Lazarovits G, Raizada MN (2014). Impact of swapping soils on the endophytic bacterial communities of pre-domesticated, ancient and modern maize. BMC Plant Biology, 14, 233. DOI: 10.1186/s12870-014-0233-3.
DOI |
| [53] |
Johnston-Monje D, Raizada MN (2011). Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLOS ONE, 6, e20396. DOI: 10.1371/journal.pone.0020396.
DOI URL |
| [54] |
Kim DH, Kaashyap M, Rathore A, Das RR, Parupalli S, Upadhyaya HD, Gopalakrishnan S, Gaur PM, Singh S, Kaur J, Yasin M, Varshney RK (2014). Phylogenetic diversity of Mesorhizobium in chickpea. Journal of Biosciences, 39, 513-517.
DOI URL |
| [55] |
Kim H, Lee KK, Jeon J, Harris WA, Lee YH (2020). Domestication of Oryza species eco-evolutionarily shapes bacterial and fungal communities in rice seed. Microbiome, 8, 20. DOI: 10.1186/s40168-020-00805-0.
DOI URL |
| [56] |
Knief C, Delmotte N, Chaffron S, Stark M, Innerebner G, Wassmann R, von Mering C, Vorholt JA (2012). Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. The ISME Journal, 6, 1378-1390.
DOI URL |
| [57] |
Kobae Y (2019). Dynamic phosphate uptake in arbuscular mycorrhizal roots under field conditions. Frontiers in Environmental Science, 6, 159. DOI: 10.3389/fenvs.2018.00159.
DOI URL |
| [58] |
Kwak MJ, Kong HG, Choi K, Kwon SK, Song JY, Lee J, Lee PA, Choi SY, Seo M, Lee HJ, Jung EJ, Park H, Roy N, Kim H, Lee MM, et al. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nature Biotechnology, 36, 1100-1109.
DOI URL |
| [59] |
Lajoie G, Maglione R, Kembel SW (2020). Adaptive matching between phyllosphere bacteria and their tree hosts in a neotropical forest. Microbiome, 8, 70. DOI: 10.1186/s40168-020-00844-7.
DOI URL |
| [60] | Leff JW, Lynch RC, Kane NC, Fierer N (2017). Plant domestication and the assembly of bacterial and fungal communities associated with strains of the common sunflower, Helianthus annuus. New phytologist, 214, 412-423. |
| [61] |
Liang JL, Liu J, Jia P, Yang TT, Zeng QW, Zhang SC, Liao B, Shu WS, Li JT (2020). Novel phosphate-solubilizing bacteria enhance soil phosphorus cycling following ecological restoration of land degraded by mining. The ISME Journal, 14, 1600-1613.
DOI URL |
| [62] |
Liu A, Ku YS, Contador CA, Lam HM (2020a). The impacts of domestication and agricultural practices on legume nutrient acquisition through symbiosis with rhizobia and arbuscular mycorrhizal fungi. Frontiers in Genetics, 11, 583954. DOI: 10.3389/fgene.2020.583954.
DOI URL |
| [63] |
Liu F, Hewezi T, Lebeis SL, Pantalone V, Grewal PS, Staton ME (2019). Soil indigenous microbiome and plant genotypes cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiology, 19, 201. DOI: 10.1186/s12866-019-1572-x.
DOI URL |
| [64] |
Liu H, Brettell LE, Qiu Z, Singh BK (2020b). Microbiome- mediated stress resistance in plants. Trends in Plant Science, 25, 733-743.
DOI URL |
| [65] |
Liu HY, Li QP, Xing YZ (2018a). Genes contributing to domestication of rice seed traits and its global expansion. Genes, 9, 489. DOI: 10.3390/genes9100489.
DOI URL |
| [66] |
Liu J, Abdelfattah A, Norelli J, Burchard E, Schena L, Droby S, Wisniewski M (2018b). Apple endophytic microbiota of different rootstock/scion combinations suggests a genotype-specific influence. Microbiome, 6, 18. DOI: 10.1186/s40168-018-0403-x.
DOI URL |
| [67] |
Liu J, Fernie AR, Yan J (2020c). The past, present, and future of maize improvement: domestication, genomics, and functional genomic routes toward crop enhancement. Plant Communications, 1, 100010. DOI: 10.1016/j.xplc.2019.100010.
DOI URL |
| [68] |
Llorens E, Sharon O, Camañes G, García-Agustín P, Sharon A (2019). Endophytes from wild cereals protect wheat plants from drought by alteration of physiological responses of the plants to water stress. Environmental Microbiology, 21, 3299-3312.
DOI URL |
| [69] |
Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL (2012). Defining the core Arabidopsis thaliana root microbiome. Nature, 488, 86-90.
DOI URL |
| [70] |
Lynch JP, Brown KM (2012). New roots for agriculture: exploiting the root phenome. Philosophical Transactions of the Royal Society B, 367, 1598-1604.
DOI URL |
| [71] |
Maignien L, DeForce EA, Chafee ME, Eren AM, Simmons SL (2014). Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. mBio, 5, e00682-13. DOI: 10.1128/mBio.00682-13.
DOI |
| [72] |
Martínez-Romero E, Aguirre-Noyola JL, Taco-Taype N, Martínez-Romero J, Zuñiga-Dávila D (2020). Plant microbiota modified by plant domestication. Systematic and Applied Microbiology, 43, 126106. DOI: 10.1016/j.syapm.2020.126106.
DOI URL |
| [73] |
Martín-Robles N, Lehmann A, Seco E, Aroca R, Rillig MC, Milla R (2018). Impacts of domestication on the arbuscular mycorrhizal symbiosis of 27 crop species. New Phytologist, 218, 322-334.
DOI PMID |
| [74] |
Massart S, Martinez-Medina M, Jijakli MH (2015). Biological control in the microbiome era: challenges and opportunities. Biological Control, 89, 98-108.
DOI URL |
| [75] |
Mendes LW, Raaijmakers JM, de Hollander M, Mendes R, Tsai SM (2018). Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. The ISME Journal, 12, 212-224.
DOI URL |
| [76] |
Meyer RS, DuVal AE, Jensen HR (2012). Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytologist, 196, 29-48.
DOI URL |
| [77] |
Meyer RS, Purugganan MD (2013). Evolution of crop species: genetics of domestication and diversification. Nature Reviews Genetics, 14, 840-852.
DOI URL |
| [78] |
Mezzasalma V, Sandionigi A, Guzzetti L, Galimberti A, Grando MS, Tardaguila J, Labra M (2018). Geographical and cultivar features differentiate grape microbiota in northern Italy and Spain vineyards. Frontiers in Microbiology, 9, 946. DOI: 10.3389/fmicb.2018.00946.
DOI PMID |
| [79] |
Milla R, Matesanz S (2017). Growing larger with domestication: a matter of physiology, morphology or allocation? Plant Biology, 19, 475-483.
DOI PMID |
| [80] |
Mina D, Pereira JA, Lino-Neto T, Baptista P (2020). Epiphytic and endophytic bacteria on olive tree phyllosphere: exploring tissue and cultivar effect. Microbial Ecology, 80, 145-157.
DOI URL |
| [81] |
Oburger E, Jones DL (2018). Sampling root exudates—Mission impossible? Rhizosphere, 6, 116-133.
DOI URL |
| [82] |
Ofek-Lalzar M, Gur Y, Ben-Moshe S, Sharon O, Kosman E, Mochli E, Sharon A (2016). Diversity of fungal endophytes in recent and ancient wheat ancestors Triticum dicoccoides and Aegilops sharonensis. FEMS Microbiology Ecology, 92, fiw152. DOI: 10.1093/femsec/fiw152.
DOI URL |
| [83] |
Özkurt E, Hassani MA, Sesiz U, Künzel S, Dagan T, Özkan H, Stukenbrock EH (2020). Seed-derived microbial colonization of wild emmer and domesticated bread wheat (Triticum dicoccoides and T. aestivum) seedlings shows pronounced differences in overall diversity and composition. mBio, 11, e02637-20. DOI: 10.1128/mBio.02637-20.
DOI |
| [84] |
Pang ZQ, Xu P, Yu DQ (2020). Environmental adaptation of the root microbiome in two rice ecotypes. Microbiological Research, 241, 126588. DOI: 10.1016/j.micres.2020.126588.
DOI URL |
| [85] |
Pascale A, Proietti S, Pantelides IS, Stringlis IA (2020). Modulation of the root microbiome by plant molecules: the basis for targeted disease suppression and plant growth promotion. Frontiers in Plant Science, 10, 1741. DOI: 10.3389/fpls.2019.01741.
DOI URL |
| [86] |
Pérez-Jaramillo JE, Carrión VJ, Bosse M, Ferrão LFV, de Hollander M, Garcia AAF, Ramírez CA, Mendes R, Raaijmakers JM (2017). Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. The ISME Journal, 11, 2244-2257.
DOI URL |
| [87] |
Pérez-Jaramillo JE, Carrión VJ, de Hollander M, Raaijmakers JM (2018). The wild side of plant microbiomes. Microbiome, 6, 143. DOI: 10.1186/s40168-018-0519-z.
DOI PMID |
| [88] |
Pérez-Jaramillo JE, de Hollander M, Ramírez CA, Mendes R, Raaijmakers JM, Carrión VJ (2019). Deciphering rhizosphere microbiome assembly of wild and modern common bean (Phaseolus vulgaris) in native and agricultural soils from Colombia. Microbiome, 7, 114. DOI: 10.1186/s40168-019-0727-1.
DOI PMID |
| [89] |
Pérez-Jaramillo JE, Mendes R, Raaijmakers JM (2016). Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Molecular Biology, 90, 635-644.
DOI PMID |
| [90] |
Popowska M, Rzeczycka M, Miernik A, Krawczyk-Balska A, Walsh F, Duffy B (2012). Influence of soil use on prevalence of tetracycline, streptomycin, and erythromycin resistance and associated resistance genes. Antimicrobial Agents and Chemotherapy, 56, 1434-1443.
DOI URL |
| [91] |
Porter SS, Sachs JL (2020). Agriculture and the disruption of plant-microbial symbiosis. Trends in Ecology & Evolution, 35, 426-439.
DOI URL |
| [92] |
Preece C, Peñuelas J (2020). A return to the wild: root exudates and food security. Trends in Plant Science, 25, 14-21.
DOI URL |
| [93] |
Putra IP, Rahayu G, Hidayat I (2015). Impact of domestication on the endophytic fungal diversity associated with wild Zingiberaceae at Mount Halimun Salak National Park. HAYATI Journal of Biosciences, 22, 157-162.
DOI URL |
| [94] |
Reinhold-Hurek B, Bünger W, Burbano CS, Sabale M, Hurek T (2015). Roots shaping their microbiome: global hotspots for microbial activity. Annual Review of Phytopathology, 53, 403-424.
DOI PMID |
| [95] |
Rho H, Hsieh M, Kandel SL, Cantillo J, Doty SL, Kim SH (2018). Do endophytes promote growth of host plants under stress? A meta-analysis on plant stress mitigation by endophytes. Microbial Ecology, 75, 407-418.
DOI URL |
| [96] |
Ritpitakphong U, Falquet L, Vimoltust A, Berger A, Métraux JP, LʼHaridon F (2016). The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytologist, 210, 1033-1043.
DOI PMID |
| [97] |
Robertson-Albertyn S, Alegria Terrazas R, Balbirnie K, Blank M, Janiak A, Szarejko I, Chmielewska B, Karcz J, Morris J, Hedley PE, George TS, Bulgarelli D (2017). Root hair mutations displace the barley rhizosphere microbiota. Frontiers in Plant Science, 8, 1094. DOI: 10.3389/fpls.2017.01094.
DOI PMID |
| [98] |
Roman-Reyna V, Pinili D, Borja FN, Quibod IL, Groen SC, Alexandrov N, Mauleon R, Oliva R (2020). Characterization of the leaf microbiome from whole-genome sequencing data of the 3000 rice genomes project. Rice, 13, 72. DOI: 10.1186/s12284-020-00432-1.
DOI PMID |
| [99] |
Rosenblueth M, Ormeño-Orrillo E, López-López A, Rogel MA, Reyes-Hernández BJ, Martínez-Romero JC, Reddy PM, Martínez-Romero E (2018). Nitrogen fixation in cereals. Frontiers in Microbiology, 9, 1794. DOI: 10.3389/fmicb.2018.01794.
DOI PMID |
| [100] |
Rosier A, Bishnoi U, Lakshmanan V, Sherrier DJ, Bais HP (2016). A perspective on inter-kingdom signaling in plant- beneficial microbe interactions. Plant Molecular Biology, 90, 537-548.
DOI URL |
| [101] |
Roucou A, Violle C, Fort F, Roumet P, Ecarnot M, Vile D (2018). Shifts in plant functional strategies over the course of wheat domestication. Journal of Applied Ecology, 55, 25-37.
DOI URL |
| [102] |
Ruan YL (2014). Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annual Review of Plant Biology, 65, 33-67.
DOI URL |
| [103] |
Saleem M, Law AD, Sahib MR, Pervaiz ZH, Zhang Q (2018). Impact of root system architecture on rhizosphere and root microbiome. Rhizosphere, 6, 47-51.
DOI URL |
| [104] |
Sangabriel-Conde W, Maldonado-Mendoza IE, Mancera-López ME, Cordero-Ramírez JD, Trejo-Aguilar D, Negrete-Yankelevich S (2015). Glomeromycota associated with Mexican native maize landraces in Los Tuxtlas, Mexico. Applied Soil Ecology, 87, 63-71.
DOI URL |
| [105] |
Sapkota R, Knorr K, Jørgensen LN, OʼHanlon KA, Nicolaisen M (2015). Host genotype is an important determinant of the cereal phyllosphere mycobiome. New Phytologist, 207, 1134-1144.
DOI URL |
| [106] |
Schardl CL, Leuchtmann A, Spiering MJ (2004). Symbioses of grasses with seedborne fungal endophytes. Annual Review of Plant Biology, 55, 315-340.
DOI URL |
| [107] |
Schmidt JE, Bowles TM, Gaudin ACM (2016). Using ancient traits to convert soil health into crop yield: impact of selection on maize root and rhizosphere function. Frontiers in Plant Science, 7, 373. DOI: 10.3389/fpls.2016.00373.
DOI |
| [108] |
Schmidt JE, Kent AD, Brisson VL, Gaudin ACM (2019). Agricultural management and plant selection interactively affect rhizosphere microbial community structure and nitrogen cycling. Microbiome, 7, 146. DOI: 10.1186/s40168-019-0756-9.
DOI URL |
| [109] |
Schmidt JE, Mazza Rodrigues JL, Brisson VL, Kent A, Gaudin ACM (2020). Impacts of directed evolution and soil management legacy on the maize rhizobiome. Soil Biology & Biochemistry, 145, 107794. DOI: 10.1016/j.soilbio.2020.107794.
DOI URL |
| [110] |
Schreiter S, Sandmann M, Smalla K, Grosch R (2014). Soil type dependent rhizosphere competence and biocontrol of two bacterial inoculant strains and their effects on the rhizosphere microbial community of field-grown lettuce. PLOS ONE, 9, e103726. DOI: 10.1371/journal.pone.0103726.
DOI URL |
| [111] |
Shade A, Jacques MA, Barret M (2017). Ecological patterns of seed microbiome diversity, transmission, and assembly. Current Opinion in Microbiology, 37, 15-22.
DOI URL |
| [112] | Shaposhnikov AI, Morgounov AI, Akin B, Makarova NM, Belimov AA, Tikhonovich IA (2016). Comparative characteristics of root systems and root exudation of synthetic, landrace and modern wheat varieties. Agricultural Biology, 51, 68-78. |
| [113] |
Shenton M, Iwamoto C, Kurata N, Ikeo K (2016). Effect of wild and cultivated rice genotypes on rhizosphere bacterial community composition. Rice, 9, 42. DOI: 10.1186/s12284-016-0111-8.
DOI |
| [114] |
Shi S, Chang J, Tian L, Nasir F, Ji L, Li X, Tian C (2019a). Comparative analysis of the rhizomicrobiome of the wild versus cultivated crop: insights from rice and soybean. Archives of Microbiology, 201, 879-888.
DOI URL |
| [115] |
Shi S, Nuccio E, Herman DJ, Rijkers R, Estera K, Li J, da Rocha UN, He Z, Pett-Ridge J, Brodie EL, Zhou J, Firestone M (2015). Successional trajectories of rhizosphere bacterial communities over consecutive seasons. mBio, 6, e00746. DOI: 10.1128/mBio.00746-15.
DOI |
| [116] |
Shi S, Richardson AE, O’Callaghan M, DeAngelis KM, Jones EE, Stewart A, Firestone MK, Condron LM (2011). Effects of selected root exudate components on soil bacterial communities. FEMS Microbiology Ecology, 77, 600-610.
DOI URL |
| [117] |
Shi S, Tian L, Nasir F, Li X, Li W, Tran LP, Tian C (2018). Impact of domestication on the evolution of rhizomicrobiome of rice in response to the presence of Magnaporthe oryzae. Plant Physiology and Biochemistry, 132, 156-165.
DOI URL |
| [118] |
Shi S, Tian L, Xu S, Ji L, Nasir F, Li X, Song Z, Tian C (2019b). The rhizomicrobiomes of wild and cultivated crops react differently to fungicides. Archives of Microbiology, 201, 477-486.
DOI URL |
| [119] |
Singh P, Santoni S, This P, Péros JP (2018). Genotype-environment interaction shapes the microbial assemblage in grapevine’s phyllosphere and carposphere: an NGS approach. Microorganisms, 6, 96. DOI: 10.3390/microorganisms6040096.
DOI URL |
| [120] |
Singh P, Santoni S, Weber A, This P, Péros JP (2019). Understanding the phyllosphere microbiome assemblage in grape species (Vitaceae) with amplicon sequence data structures. Scientific Reports, 9, 14294. DOI: 10.1038/s41598-019-50839-0.
DOI URL |
| [121] |
Spor A, Roucou A, Mounier A, Bru D, Breuil MC, Fort F, Vile D, Roumet P, Philippot L, Violle C (2020). Domestication-driven changes in plant traits associated with changes in the assembly of the rhizosphere microbiota in tetraploid wheat. Scientific Reports, 10, 12234. DOI: 10.1038/s41598-020-69175-9.
DOI URL |
| [122] |
Stenberg JA, Heil M, Åhman I, Björkman C (2015). Optimizing crops for biocontrol of pests and disease. Trends in Plant Science, 20, 698-712.
DOI PMID |
| [123] |
Stitzer MC, Ross-Ibarra J (2018). Maize domestication and gene interaction. New Phytologist, 220, 395-408.
DOI URL |
| [124] | Stringlis IA, Yu K, Feussner K, de Jonge R, van Bentum S, van Verk MC, Berendsen RL, Bakker PAHM, Feussner I, Pieterse CMJ (2018). MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proceedings of the National Academy of Sciences of the United States of America, 115, E5213-E5222. |
| [125] |
Szczepaniec A, Widney SE, Bernal JS, Eubanks MD (2013). Higher expression of induced defenses in teosintes (Zea spp.) is correlated with greater resistance to fall armyworm, Spodoptera frugiperda. Entomologia Experimentalis et Applicata, 146, 242-251.
DOI URL |
| [126] |
Szoboszlay M, Lambers J, Chappell J, Kupper JV, Moe LA, McNear Jr DH (2015). Comparison of root system architecture and rhizosphere microbial communities of Balsas teosinte and domesticated corn cultivars. Soil Biology & Biochemistry, 80, 34-44.
DOI URL |
| [127] |
Tan S, Liu J, Fang Y, Hedlund BP, Lian ZH, Huang LY, Li JT, Huang LN, Li WJ, Jiang HC, Dong HL, Shu WS (2019). Insights into ecological role of a new deltaproteobacterial order Candidatus Acidulodesulfobacterales by metagenomics and metatranscriptomics. The ISME Journal, 13, 2044-2057.
DOI URL |
| [128] |
Tian L, Lin X, Tian J, Ji L, Chen Y, Tran LP, Tian C (2020a). Research advances of beneficial microbiota associated with crop plants. International Journal of Molecular Sciences, 21, 1792. DOI: 10.3390/ijms21051792.
DOI URL |
| [129] |
Tian L, Shi S, Ma L, Tran LSP, Tian C (2020b). Community structures of the rhizomicrobiomes of cultivated and wild soybeans in their continuous cropping. Microbiological Research, 232, 126390. DOI: 10.1016/j.micres.2019.126390.
DOI URL |
| [130] |
Tkacz A, Pini F, Turner TR, Bestion E, Simmonds J, Howell P, Greenland A, Cheema J, Emms DM, Uauy C, Poole PS (2020). Agricultural selection of wheat has been shaped by plant-microbe interactions. Frontiers in Microbiology, 11, 132. DOI: 10.3389/fmicb.2020.00132.
DOI URL |
| [131] |
Truyens S, Weyens N, Cuypers A, Vangronsveld J (2015). Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environmental Microbiology Reports, 7, 40-50.
DOI URL |
| [132] |
Vacher C, Hampe A, Porté AJ, Sauer U, Compant S, Morris CE (2016). The phyllosphere: microbial jungle at the plant- climate interface. Annual Review of Ecology, Evolution and Systematics, 47, 1-24.
DOI URL |
| [133] |
van der Heijden MGA, Schlaeppi K (2015). Root surface as a frontier for plant microbiome research. Proceedings of the National Academy of Sciences of the United States of America, 112, 2299-2300.
DOI PMID |
| [134] |
van Deynze A, Zamora P, Delaux PM, Heitmann C, Jayaraman D, Rajasekar S, Graham D, Maeda J, Gibson D, Schwartz KD, Berry AM, Bhatnagar S, Jospin G, Darling A, Jeannotte R, et al. (2018). Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLOS Biology, 16, e2006352. DOI: 10.1371/journal.pbio.2006352.
DOI URL |
| [135] |
Vandenkoornhuyse P, Quaiser A, Duhamel M, van AL, Dufresne A (2015). The importance of the microbiome of the plant holobiont. New phytologist, 206, 1196-1206.
DOI PMID |
| [136] |
Wagner MR, Busby PE, Balint-Kurti P (2020a). Analysis of leaf microbiome composition of near-isogenic maize lines differing in broad-pectrum disease resistance. New Phytologist, 225, 2152-2165.
DOI URL |
| [137] |
Wagner MR, Lundberg DS, del Rio TG, Tringe SG, Dangl JL, Mitchell-Olds T (2016). Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nature Communications, 7, 12151. DOI: 10.1038/ncomms12151.
DOI URL |
| [138] |
Wagner MR, Roberts JH, Balint-Kurti P, Holland JB (2020b). Heterosis of leaf and rhizosphere microbiomes in field-grown maize. New Phytologist, 228, 1055-1069.
DOI URL |
| [139] |
Wallace JG, Kremling KA, Kovar LL, Buckler ES (2018). Quantitative genetics of the maize leaf microbiome. Phytobiomes Journal, 2, 208-224.
DOI URL |
| [140] |
Wang YT, Li T, Li Y, Björn LO, Rosendahl S, Olsson PA, Li SS, Fu XL (2015). Community dynamics of arbuscular mycorrhizal fungi in high-input and intensively irrigated rice cultivation systems. Applied and Environmental Microbiology, 81, 2958-2965.
DOI URL |
| [141] |
Wang YT, Li YW, Bao XZ, Björn LO, Li SS, Olsson PA (2016). Response differences of arbuscular mycorrhizal fungi communities in the roots of an aquatic and a semiaquatic species to various flooding regimes. Plant and Soil, 403, 361-373.
DOI URL |
| [142] |
Wang YT, Li YW, Li SS, Rosendahl S (2021). Ignored diversity of arbuscular mycorrhizal fungi in co-occurring mycotrophic and non-mycotrophic plants. Mycorrhiza, 31, 93-102.
DOI URL |
| [143] |
Wei N, Ashman TL (2018). The effects of host species and sexual dimorphism differ among root, leaf and flower microbiomes of wild strawberries in situ. Scientific Reports, 8, 5195. DOI: 10.1038/s41598-018-23518-9.
DOI PMID |
| [144] |
White JF, Kingsley KL, Zhang Q, Verma R, Obi N, Dvinskikh S, Elmore MT, Verma SK, Gond SK, Kowalski KP (2019). Review: Endophytic microbes and their potential applications in crop management. Pest Management Science, 75, 2558-2565.
DOI URL |
| [145] |
Whitehead SR, Turcotte MM, Poveda K (2017). Domestication impacts on plant-herbivore interactions: a meta-analysis. Philosophical Transactions of the Royal Society B, 372, 20160034. DOI: 10.1098/rstb.2016.0034.
DOI URL |
| [146] | Xing X, Koch AMP, Jones AM, Ragone D, Murch S, Hart MM (2012). Mutualism breakdown in breadfruit domestication. Proceedings of the Royal Society B, 279, 1122-1130. |
| [147] | Xiong C, Zhu YG, Wang JT, Singh B, Han LL, Shen JP, Li PP, Wang GB, Wu CF, Ge AH, Zhang LM, He JZ (2021). Host selection shapes crop microbiome assembly and network complexity. New Phytologist, 229, 1091-1104. |
| [148] |
Yoneyama K (2019). How do strigolactones ameliorate nutrient deficiencies in plants? Cold Spring Harbor Perspectives in Biology, 11, a034686. DOI: 10.1101/cshperspect.a034686.
DOI URL |
| [149] | Yu K, Pieterse CMJ, Bakker PAHM, Berendsen RL (2019). Beneficial microbes going underground of root immunity. Plant, Cell & Environment, 42, 2860-2870. |
| [150] |
Zachow C, Müller H, Tilcher R, Berg G (2014). Differences between the rhizosphere microbiome of Beta vulgaris ssp. maritima—ancestor of all beet crops—and modern sugar beets. Frontiers in Microbiology, 5, 415. DOI: 10.3389/fmicb.2014.00415.
DOI |
| [151] |
Zhang J, Liu YX, Guo X, Qin Y, Garrido-Oter R, Schulze-Lefert P, Bai Y (2021). High-throughput cultivation and identification of bacteria from the plant root microbiota. Nature Protocols, 16, 988-1012.
DOI URL |
| [152] |
Zhang SW, Wang Y, Chen X, Cui BJ, Bai ZH, Zhuang GQ (2020). Variety features differentiate microbiota in the grape leaves. Canadian Journal of Microbiology, 66, 653-663.
DOI URL |
| [153] |
Zheng YF, Liang J, Zhao DL, Meng C, Xu ZC, Xie ZH, Zhang CS (2020). The root nodule microbiome of cultivated and wild halophytic legumes showed similar diversity but distinct community structure in Yellow River Delta saline soils. Microorganisms, 8, 207. DOI: 10.3390/ microorganisms8020207.
DOI URL |
| [154] |
Zhou WH, Wang YT, Lian ZH, Yang TT, Zeng QW, Feng SW, Fang Z, Shu WS, Huang LN, Ye ZH, Liao B, Li JT (2020). Revegetation approach and plant identity unequally affect structure, ecological network and function of soil microbial community in a highly acidified mine tailings pond. Science of the Total Environment, 744, 140793. DOI: 10.1016/j.scitotenv.2020.140793.
DOI URL |
| [1] | 刘瑶 钟全林 徐朝斌 程栋梁 郑跃芳 邹宇星 张雪 郑新杰 周云若. 不同大小刨花楠细根功能性状与根际微环境关系[J]. 植物生态学报, 2024, 48(预发表): 0-0. |
| [2] | 陈科宇 邢森 唐玉 孙佳慧 任世杰 张静 纪宝明. 不同草地型土壤丛枝菌根真菌群落特征及其驱动因素[J]. 植物生态学报, 2024, 48(5): 660-674. |
| [3] | 徐子怡 金光泽. 阔叶红松林不同菌根类型幼苗细根功能性状的变异与权衡[J]. 植物生态学报, 2024, 48(5): 612-622. |
| [4] | 胡蝶 蒋欣琪 戴志聪 陈戴一 张雨 祁珊珊 杜道林. 丛枝菌根真菌提高入侵杂草南美蟛蜞菊对除草剂的耐受性[J]. 植物生态学报, 2024, 48(5): 651-659. |
| [5] | 杨安娜, 李曾燕, 牟凌, 杨柏钰, 赛碧乐, 张立, 张增可, 王万胜, 杜运才, 由文辉, 阎恩荣. 上海大金山岛不同植被类型土壤细菌群落的变异[J]. 植物生态学报, 2024, 48(3): 377-389. |
| [6] | 牛一迪, 蔡体久. 大兴安岭北部次生林演替过程中物种多样性的变化及其影响因子[J]. 植物生态学报, 2024, 48(3): 349-363. |
| [7] | 程可心, 杜尧, 李凯航, 王浩臣, 杨艳, 金一, 何晓青. 玉米与叶际微生物组的互作遗传机制[J]. 植物生态学报, 2024, 48(2): 215-228. |
| [8] | 陈昭铨, 王明慧, 胡子涵, 郎学东, 何云琼, 刘万德. 云南普洱季风常绿阔叶林幼苗的群落构建机制[J]. 植物生态学报, 2024, 48(1): 68-79. |
| [9] | 陈保冬, 付伟, 伍松林, 朱永官. 菌根真菌在陆地生态系统碳循环中的作用[J]. 植物生态学报, 2024, 48(1): 1-20. |
| [10] | 李娜, 唐士明, 郭建英, 田茹, 王姗, 胡冰, 罗永红, 徐柱文. 放牧对内蒙古草地植物群落特征影响的meta分析[J]. 植物生态学报, 2023, 47(9): 1256-1269. |
| [11] | 任悦, 高广磊, 丁国栋, 张英, 赵珮杉, 柳叶. 不同生长期樟子松外生菌根真菌群落物种组成及其驱动因素[J]. 植物生态学报, 2023, 47(9): 1298-1309. |
| [12] | 杨鑫, 任明迅. 环南海区域红树物种多样性分布格局及其形成机制[J]. 植物生态学报, 2023, 47(8): 1105-1115. |
| [13] | 于笑, 纪若璇, 任天梦, 夏新莉, 尹伟伦, 刘超. 中国北方蒙古莸群落的分布、特征和分类[J]. 植物生态学报, 2023, 47(8): 1182-1192. |
| [14] | 张中扬, 宋希强, 任明迅, 张哲. 附生维管植物生境营建作用的生态学功能[J]. 植物生态学报, 2023, 47(7): 895-911. |
| [15] | 张仲富, 王四海, 杨卫, 陈剑. 蒜头果根际细菌群落结构与功能特征对其健康状态的响应[J]. 植物生态学报, 2023, 47(7): 1020-1031. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
Copyright © 2022 版权所有 《植物生态学报》编辑部
地址: 北京香山南辛村20号, 邮编: 100093
Tel.: 010-62836134, 62836138; Fax: 010-82599431; E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
备案号: 京ICP备16067583号-19