

植物生态学报 ›› 2011, Vol. 35 ›› Issue (3): 330-336.DOI: 10.3724/SP.J.1258.2011.00330
所属专题: 生物多样性
收稿日期:2010-06-04
接受日期:2010-11-22
出版日期:2011-06-04
发布日期:2011-03-02
通讯作者:
胡庭兴
作者简介:*E-mail: hutx001@yahoo.com.cn
ZHANG Wei1,2, LUO Jian-Xun2, GU Yun-Jie2, HU Ting-Xing1,*( )
)
Received:2010-06-04
Accepted:2010-11-22
Online:2011-06-04
Published:2011-03-02
Contact:
HU Ting-Xing
摘要:
为了揭示麻疯树(Jatropha curcas)天然种群的遗传多样性和遗传结构, 采用聚苯烯酰胺凝胶垂直平板电泳技术, 对采自四川、云南、贵州3个省的10个麻疯树天然种群的叶片样本进行了同工酶分析。7个酶系统10个位点的检测结果表明: 麻疯树种群水平上的遗传多样性较高, 每位点平均等位基因数为2.428 6, 多态位点百分率为97.14%, 平均期望杂合度为0.396 4。种群间遗传分化系数为0.041 3, 种群间总的基因流较高, 为5.808 9, 种群间遗传一致度较高(Shannon信息指数为0.921 7- 0.995 3)。非加权类平均法(UPGMA)聚类结果显示, 10个种群的遗传距离与地理距离相关性不显著。麻疯树天然种群具有较低程度的遗传分化、较高的基因流, 种内及种群内多样性丰富, 这为麻疯树优良品种的选育提供了良好的遗传基础。
张炜, 罗建勋, 辜云杰, 胡庭兴. 西南地区麻疯树天然种群遗传多样性的等位酶变异. 植物生态学报, 2011, 35(3): 330-336. DOI: 10.3724/SP.J.1258.2011.00330
ZHANG Wei, LUO Jian-Xun, GU Yun-Jie, HU Ting-Xing. Allozyme variation of genetic diversity in natural populations of Jatropha curcas germplasm from different areas in southwest China. Chinese Journal of Plant Ecology, 2011, 35(3): 330-336. DOI: 10.3724/SP.J.1258.2011.00330
| 种群代号 Population code | 采样地 Seed collection site | 经纬度 Longitude and latitude | 海拔 Altitude (m) | 水系 River system | 家系数量 No. of families |
|---|---|---|---|---|---|
| LS | 卢水 Lushui | 98°08′ E, 25°08′ N | 850 | 怒江 Nujiang River | 30 |
| SB | 双柏 Shuangbo | 101°51′ E, 25°49′ N | 1 066 | 红河 Honghe River | 30 |
| YNPE | 普洱 Puer | 100°59′ E, 22°42′ N | 1 300 | 澜沧江 Lancang River | 30 |
| XSBN | 西双版纳 Xishuangbanna | 100°80′ E, 22°35′ N | 1 000 | 澜沧江 Lancang River | 30 |
| PJ | 坡脚 Pojiao | 102°55′ E, 26°74′ N | 1 080 | 金沙江 Jinsha River | 30 |
| FY | 泸西 Luxi | 103°76′ E, 24°52′N | 1 100 | 南盘江 Nanpan River | 30 |
| LB | 雷波 Leibo | 103°62′ E, 28°21′ N | 960 | 金沙江 Jinsha River | 30 |
| HPZ | 花棚子 Huapengzi | 101°55′ E, 26°22′ N | 983 | 金沙江 Jinsha River | 30 |
| HL | 会理 Huili | 101°55′ E, 26°10′ N | 943 | 金沙江 Jinsha River | 30 |
| YL | 金河 Jinhe | 101°51′ E, 27°42′ N | 1 200 | 雅砻江 Yagong River | 30 |
表1 麻疯树采样种群的地理位置和样本量
Table 1 Geographic locations and sample size of Jatropha curcas populations
| 种群代号 Population code | 采样地 Seed collection site | 经纬度 Longitude and latitude | 海拔 Altitude (m) | 水系 River system | 家系数量 No. of families |
|---|---|---|---|---|---|
| LS | 卢水 Lushui | 98°08′ E, 25°08′ N | 850 | 怒江 Nujiang River | 30 |
| SB | 双柏 Shuangbo | 101°51′ E, 25°49′ N | 1 066 | 红河 Honghe River | 30 |
| YNPE | 普洱 Puer | 100°59′ E, 22°42′ N | 1 300 | 澜沧江 Lancang River | 30 |
| XSBN | 西双版纳 Xishuangbanna | 100°80′ E, 22°35′ N | 1 000 | 澜沧江 Lancang River | 30 |
| PJ | 坡脚 Pojiao | 102°55′ E, 26°74′ N | 1 080 | 金沙江 Jinsha River | 30 |
| FY | 泸西 Luxi | 103°76′ E, 24°52′N | 1 100 | 南盘江 Nanpan River | 30 |
| LB | 雷波 Leibo | 103°62′ E, 28°21′ N | 960 | 金沙江 Jinsha River | 30 |
| HPZ | 花棚子 Huapengzi | 101°55′ E, 26°22′ N | 983 | 金沙江 Jinsha River | 30 |
| HL | 会理 Huili | 101°55′ E, 26°10′ N | 943 | 金沙江 Jinsha River | 30 |
| YL | 金河 Jinhe | 101°51′ E, 27°42′ N | 1 200 | 雅砻江 Yagong River | 30 |
| 酶系统 Enzyme system | 缩写 Abbreviation | 标准代号 EC No. | 位点数 No. of loci |
|---|---|---|---|
| 谷草转氨酶 Glutamic-oxalacetic transaminase | GOT | (E.C.2.6.1.1) | 2 |
| 莽草酸脱氢酶 Shikimate acid dehydrogenase | SHDH | (E.C.1.1.1.25) | 1 |
| 6-磷酸葡萄糖脱氢酶 Phosphoglucose dehydrogenase | 6PGD | (E.C 1.1.1.44) | 2 |
| 丙氨酸氨基肽酶 Alanine aminopeptidase | AAP | (E.C.3.4.11. I) | 2 |
| 磷酸葡萄糖变位酶 Phosphogluco mutase | PGM | (E.C.5.4.2.23) | 3 |
| 磷酸葡萄糖异构酶 Phosphogluco isomerase | PGI | (E.C.5.3.1.9) | 4 |
| 甲基萘醌还原酶 Menadione reductase | MNR | (E.C.1.6.99.2) | 7 |
表2 7种酶系统和每个酶系统检测到的位点数
Table 2 Seven enzyme systems assayed and No. of loci detected in each enzyme system
| 酶系统 Enzyme system | 缩写 Abbreviation | 标准代号 EC No. | 位点数 No. of loci |
|---|---|---|---|
| 谷草转氨酶 Glutamic-oxalacetic transaminase | GOT | (E.C.2.6.1.1) | 2 |
| 莽草酸脱氢酶 Shikimate acid dehydrogenase | SHDH | (E.C.1.1.1.25) | 1 |
| 6-磷酸葡萄糖脱氢酶 Phosphoglucose dehydrogenase | 6PGD | (E.C 1.1.1.44) | 2 |
| 丙氨酸氨基肽酶 Alanine aminopeptidase | AAP | (E.C.3.4.11. I) | 2 |
| 磷酸葡萄糖变位酶 Phosphogluco mutase | PGM | (E.C.5.4.2.23) | 3 |
| 磷酸葡萄糖异构酶 Phosphogluco isomerase | PGI | (E.C.5.3.1.9) | 4 |
| 甲基萘醌还原酶 Menadione reductase | MNR | (E.C.1.6.99.2) | 7 |
| 位点 Locus | 等位基因 Allele | 种群 Population | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 金河 YL | 坡脚 PJ | 花棚子 HPZ | 雷波 LB | 法依 FY | 双柏 SB | 版纳 XSBN | 普洱 YNPE | 会理 HL | 卢水 LS | 平均 Mean | ||
| AAP-1 | A | 0.725 | 0.675 | 0.650 | 0.750 | 0500 | 0.675 | 0.600 | 0.750 | 0.550 | 0.650 | 0.652 |
| B | 0.275 | 0.325 | 0.350 | 0.250 | 0.500 | 0.325 | 0.400 | 0.250 | 0.450 | 0.350 | 0.348 | |
| SHDH | A | 0.300 | 0.350 | 0.300 | 0.600 | 0.600 | 0.775 | 0.600 | 0.500 | 0.525 | 0.325 | 0.487 |
| B | 0.675 | 0.500 | 0.500 | 0.400 | 0.400 | 0.175 | 0.400 | 0.500 | 0.450 | 0.675 | 0.467 | |
| C | 0.025 | 0.150 | 0.200 | 0.000 | 0.000 | 0.050 | 0.000 | 0.000 | 0.025 | 0.000 | 0.045 | |
| GOT-2 | A | 0.425 | 0.375 | 0.325 | 0.350 | 0.350 | 0.350 | 0.300 | 0.400 | 0.325 | 0.275 | 0.347 |
| B | 0.575 | 0.625 | 0.675 | 0.650 | 0.650 | 0.650 | 0.700 | 0.600 | 0.625 | 0.725 | 0.647 | |
| PGI-4 | A | 0.625 | 0.500 | 0.450 | 0.550 | 0.475 | 0.525 | 0.575 | 0.500 | 0.550 | 0.550 | 0.530 |
| B | 0.375 | 0.500 | 0.500 | 0.375 | 0.475 | 0.350 | 0.425 | 0.500 | 0.450 | 0.450 | 0.440 | |
| C | 0.000 | 0.000 | 0.050 | 0.075 | 0.050 | 0.125 | 0.000 | 0.000 | 0.000 | 0.000 | 0.030 | |
| 6PGD-1 | A | 1.000 | 0.600 | 0.700 | 0.875 | 0.875 | 0.750 | 0.650 | 0.800 | 0.750 | 1.000 | 0.800 |
| B | 0.000 | 0.400 | 0.300 | 0.125 | 0.125 | 0.250 | 0.350 | 0.200 | 0.250 | 0.000 | 0.200 | |
| MNR-3 | A | 0. 250 | 0.175 | 0.175 | 0.200 | 0.175 | 0.150 | 0.200 | 0.275 | 0.250 | 0.175 | 0.177 |
| B | 0.750 | 0.825 | 0.825 | 0.800 | 0.825 | 0.850 | 0.800 | 0.725 | 0.750 | 0.825 | 0.797 | |
| PGM-2 | A | 0.700 | 0.875 | 0.850 | 0.700 | 0.825 | 0.650 | 0.700 | 0.625 | 0.575 | 0.675 | 0.718 |
| B | 0.300 | 0.125 | 0.150 | 0.300 | 0.175 | 0.325 | 0.300 | 0.375 | 0.425 | 0.325 | 0.280 | |
| C | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.025 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | |
| 基因频率平均值 Mean value of gene frequency | 0.397 | 0.412 | 0.412 | 0.412 | 0.412 | 0.412 | 0.412 | 0.412 | 0.409 | 0.412 | 0.410 | |
| 等位基因数 Number of alleles | 14 | 15 | 16 | 15 | 15 | 17 | 15 | 14 | 15 | 13 | 14.9 | |
表3 10个麻疯树种群等位基因的频率分布
Table 3 Allele frequencies in ten natural populations of Jatropha curcas
| 位点 Locus | 等位基因 Allele | 种群 Population | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 金河 YL | 坡脚 PJ | 花棚子 HPZ | 雷波 LB | 法依 FY | 双柏 SB | 版纳 XSBN | 普洱 YNPE | 会理 HL | 卢水 LS | 平均 Mean | ||
| AAP-1 | A | 0.725 | 0.675 | 0.650 | 0.750 | 0500 | 0.675 | 0.600 | 0.750 | 0.550 | 0.650 | 0.652 |
| B | 0.275 | 0.325 | 0.350 | 0.250 | 0.500 | 0.325 | 0.400 | 0.250 | 0.450 | 0.350 | 0.348 | |
| SHDH | A | 0.300 | 0.350 | 0.300 | 0.600 | 0.600 | 0.775 | 0.600 | 0.500 | 0.525 | 0.325 | 0.487 |
| B | 0.675 | 0.500 | 0.500 | 0.400 | 0.400 | 0.175 | 0.400 | 0.500 | 0.450 | 0.675 | 0.467 | |
| C | 0.025 | 0.150 | 0.200 | 0.000 | 0.000 | 0.050 | 0.000 | 0.000 | 0.025 | 0.000 | 0.045 | |
| GOT-2 | A | 0.425 | 0.375 | 0.325 | 0.350 | 0.350 | 0.350 | 0.300 | 0.400 | 0.325 | 0.275 | 0.347 |
| B | 0.575 | 0.625 | 0.675 | 0.650 | 0.650 | 0.650 | 0.700 | 0.600 | 0.625 | 0.725 | 0.647 | |
| PGI-4 | A | 0.625 | 0.500 | 0.450 | 0.550 | 0.475 | 0.525 | 0.575 | 0.500 | 0.550 | 0.550 | 0.530 |
| B | 0.375 | 0.500 | 0.500 | 0.375 | 0.475 | 0.350 | 0.425 | 0.500 | 0.450 | 0.450 | 0.440 | |
| C | 0.000 | 0.000 | 0.050 | 0.075 | 0.050 | 0.125 | 0.000 | 0.000 | 0.000 | 0.000 | 0.030 | |
| 6PGD-1 | A | 1.000 | 0.600 | 0.700 | 0.875 | 0.875 | 0.750 | 0.650 | 0.800 | 0.750 | 1.000 | 0.800 |
| B | 0.000 | 0.400 | 0.300 | 0.125 | 0.125 | 0.250 | 0.350 | 0.200 | 0.250 | 0.000 | 0.200 | |
| MNR-3 | A | 0. 250 | 0.175 | 0.175 | 0.200 | 0.175 | 0.150 | 0.200 | 0.275 | 0.250 | 0.175 | 0.177 |
| B | 0.750 | 0.825 | 0.825 | 0.800 | 0.825 | 0.850 | 0.800 | 0.725 | 0.750 | 0.825 | 0.797 | |
| PGM-2 | A | 0.700 | 0.875 | 0.850 | 0.700 | 0.825 | 0.650 | 0.700 | 0.625 | 0.575 | 0.675 | 0.718 |
| B | 0.300 | 0.125 | 0.150 | 0.300 | 0.175 | 0.325 | 0.300 | 0.375 | 0.425 | 0.325 | 0.280 | |
| C | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.025 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | |
| 基因频率平均值 Mean value of gene frequency | 0.397 | 0.412 | 0.412 | 0.412 | 0.412 | 0.412 | 0.412 | 0.412 | 0.409 | 0.412 | 0.410 | |
| 等位基因数 Number of alleles | 14 | 15 | 16 | 15 | 15 | 17 | 15 | 14 | 15 | 13 | 14.9 | |
| 种群 Population | 个体 Individual | 等位基因数 Number of alleles | 多态位点百分率 P | Shannon信息指数 I | 观测杂合度 Ho | 期望杂合度 He |
|---|---|---|---|---|---|---|
| LS | 30 | 2.0 (0.2) | 85.7 | 0.546 2 | 0.493 (0.092) | 0.382 (0.065) |
| SB | 30 | 2.1 (0.1) | 100.0 | 0.642 5 | 0.600 (0.080) | 0.440 (0.051) |
| YNPE | 30 | 2.3 (0.2) | 100.0 | 0.665 8 | 0.600 (0.076) | 0.443 (0.050) |
| XSBN | 30 | 2.1 (0.1) | 100.0 | 0.608 8 | 0.564 (0.090) | 0.413 (0.043) |
| PJ | 30 | 2.1 (0.1) | 100.0 | 0.596 4 | 0.529 (0.091) | 0.407 (0.050) |
| FY | 30 | 2.4 (0.2) | 100.0 | 0.659 8 | 0.550 (0.089) | 0.432 (0.040) |
| LB | 30 | 2.0 ( 0.0) | 100.0 | 0.628 2 | 0.679 (0.060) | 0.449 (0.023) |
| HPZ | 30 | 2.0 (0.0) | 100.0 | 0.624 5 | 0.629 (0.055) | 0.446 (0.027) |
| HL | 30 | 2.1 (0.1) | 100.0 | 0.662 0 | 0.679 (0.068) | 0.472 (0.023) |
| YL | 30 | 1.9 ( 0.1) | 85.7 | 0.521 2 | 0.471 (0.091) | 0.368 (0.066) |
| 平均 | 30 | 2.1 (0.1) | 97.1 | 0.615 5 | 0.579 (0.079) | 0.425 (0.043) |
表4 10个麻疯树天然种群的遗传多样性
Table 4 Genetic diversity in ten natural populations of Jatropha curcas
| 种群 Population | 个体 Individual | 等位基因数 Number of alleles | 多态位点百分率 P | Shannon信息指数 I | 观测杂合度 Ho | 期望杂合度 He |
|---|---|---|---|---|---|---|
| LS | 30 | 2.0 (0.2) | 85.7 | 0.546 2 | 0.493 (0.092) | 0.382 (0.065) |
| SB | 30 | 2.1 (0.1) | 100.0 | 0.642 5 | 0.600 (0.080) | 0.440 (0.051) |
| YNPE | 30 | 2.3 (0.2) | 100.0 | 0.665 8 | 0.600 (0.076) | 0.443 (0.050) |
| XSBN | 30 | 2.1 (0.1) | 100.0 | 0.608 8 | 0.564 (0.090) | 0.413 (0.043) |
| PJ | 30 | 2.1 (0.1) | 100.0 | 0.596 4 | 0.529 (0.091) | 0.407 (0.050) |
| FY | 30 | 2.4 (0.2) | 100.0 | 0.659 8 | 0.550 (0.089) | 0.432 (0.040) |
| LB | 30 | 2.0 ( 0.0) | 100.0 | 0.628 2 | 0.679 (0.060) | 0.449 (0.023) |
| HPZ | 30 | 2.0 (0.0) | 100.0 | 0.624 5 | 0.629 (0.055) | 0.446 (0.027) |
| HL | 30 | 2.1 (0.1) | 100.0 | 0.662 0 | 0.679 (0.068) | 0.472 (0.023) |
| YL | 30 | 1.9 ( 0.1) | 85.7 | 0.521 2 | 0.471 (0.091) | 0.368 (0.066) |
| 平均 | 30 | 2.1 (0.1) | 97.1 | 0.615 5 | 0.579 (0.079) | 0.425 (0.043) |
| 位点 Locus | 杂合度 Heterozygosity | 种群 Population | 平均值 Mean | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 金河 YL | 坡脚 PJ | 花棚子 HPZ | 雷波 LB | 法依 FY | 双柏 SB | 版纳 XSBN | 普洱 YNPE | 会理 HL | 卢水 LS | |||
| AAP-1 | Ho | 0.450 | 0.350 | 0.300 | 0.500 | 0.400 | 0.350 | 0.200 | 0.500 | 0.100 | 0.400 | 0.355 |
| He | 0.550 | 0.650 | 0.700 | 0.500 | 0.600 | 0.650 | 0.800 | 0.500 | 0.900 | 0.600 | 0.645 | |
| SHDH | Ho | 0.600 | 0.300 | 0.200 | 0.200 | 0.200 | 0.550 | 0.200 | 0.300 | 0.150 | 0.550 | 0.325 |
| He | 0.400 | 0.700 | 0.800 | 0.800 | 0.800 | 0.450 | 0.800 | 0.700 | 0.850 | 0.450 | 0.675 | |
| GOT-2 | Ho | 0.350 | 0.250 | 0.350 | 0.300 | 0.300 | 0.300 | 0.400 | 0.200 | 0.250 | 0.450 | 0.315 |
| He | 0.650 | 0.750 | 0.650 | 0.700 | 0.700 | 0.700 | 0.600 | 0.800 | 0.750 | 0.550 | 0.685 | |
| PGI-4 | Ho | 0.250 | 0.300 | 0.200 | 0.100 | 0.250 | 0.500 | 0.150 | 0.300 | 0.300 | 0.300 | 0.265 |
| He | 0.750 | 0.700 | 0.800 | 0.900 | 0.050 | 0.950 | 0.850 | 0.700 | 0.700 | 0.700 | 0.710 | |
| 6PGD-1 | Ho | 1.000 | 0.200 | 0.400 | 0.750 | 0.750 | 0.500 | 0.300 | 0.600 | 0.500 | 1.000 | 0.600 |
| He | 0.000 | 0.800 | 0.600 | 0.250 | 0.250 | 0.500 | 0.700 | 0.400 | 0.500 | 0.000 | 0.400 | |
| MNR-3 | Ho | 0.500 | 0.650 | 0.650 | 0.600 | 0.650 | 0.700 | 0.600 | 0.450 | 0.600 | 0.650 | 0.605 |
| He | 0.500 | 0.350 | 0.350 | 0.400 | 0.350 | 0.300 | 0.400 | 0.550 | 0.400 | 0.350 | 0.395 | |
| PGM-2 | Ho | 0.400 | 0.750 | 0.700 | 0.600 | 0.750 | 0.700 | 0.400 | 0.250 | 0.350 | 0.350 | 0.525 |
| He | 0.600 | 0.250 | 0.300 | 0.400 | 0.250 | 0.300 | 0.600 | 0.750 | 0.650 | 0.650 | 0.475 | |
| 平均值 Mean | Ho | 0.507 | 0.400 | 0.400 | 0.436 | 0.471 | 0.514 | 0.321 | 0.371 | 0.321 | 0.528 | 0.427 |
| He | 0.493 | 0.600 | 0.600 | 0.564 | 0.429 | 0.550 | 0.679 | 0.628 | 0.678 | 0.471 | 0.569 | |
表5 麻疯树10个种群的单位点杂合度
Table 5 Observed heterozygosity at single locus detected in ten populations of Jatropha curcas
| 位点 Locus | 杂合度 Heterozygosity | 种群 Population | 平均值 Mean | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 金河 YL | 坡脚 PJ | 花棚子 HPZ | 雷波 LB | 法依 FY | 双柏 SB | 版纳 XSBN | 普洱 YNPE | 会理 HL | 卢水 LS | |||
| AAP-1 | Ho | 0.450 | 0.350 | 0.300 | 0.500 | 0.400 | 0.350 | 0.200 | 0.500 | 0.100 | 0.400 | 0.355 |
| He | 0.550 | 0.650 | 0.700 | 0.500 | 0.600 | 0.650 | 0.800 | 0.500 | 0.900 | 0.600 | 0.645 | |
| SHDH | Ho | 0.600 | 0.300 | 0.200 | 0.200 | 0.200 | 0.550 | 0.200 | 0.300 | 0.150 | 0.550 | 0.325 |
| He | 0.400 | 0.700 | 0.800 | 0.800 | 0.800 | 0.450 | 0.800 | 0.700 | 0.850 | 0.450 | 0.675 | |
| GOT-2 | Ho | 0.350 | 0.250 | 0.350 | 0.300 | 0.300 | 0.300 | 0.400 | 0.200 | 0.250 | 0.450 | 0.315 |
| He | 0.650 | 0.750 | 0.650 | 0.700 | 0.700 | 0.700 | 0.600 | 0.800 | 0.750 | 0.550 | 0.685 | |
| PGI-4 | Ho | 0.250 | 0.300 | 0.200 | 0.100 | 0.250 | 0.500 | 0.150 | 0.300 | 0.300 | 0.300 | 0.265 |
| He | 0.750 | 0.700 | 0.800 | 0.900 | 0.050 | 0.950 | 0.850 | 0.700 | 0.700 | 0.700 | 0.710 | |
| 6PGD-1 | Ho | 1.000 | 0.200 | 0.400 | 0.750 | 0.750 | 0.500 | 0.300 | 0.600 | 0.500 | 1.000 | 0.600 |
| He | 0.000 | 0.800 | 0.600 | 0.250 | 0.250 | 0.500 | 0.700 | 0.400 | 0.500 | 0.000 | 0.400 | |
| MNR-3 | Ho | 0.500 | 0.650 | 0.650 | 0.600 | 0.650 | 0.700 | 0.600 | 0.450 | 0.600 | 0.650 | 0.605 |
| He | 0.500 | 0.350 | 0.350 | 0.400 | 0.350 | 0.300 | 0.400 | 0.550 | 0.400 | 0.350 | 0.395 | |
| PGM-2 | Ho | 0.400 | 0.750 | 0.700 | 0.600 | 0.750 | 0.700 | 0.400 | 0.250 | 0.350 | 0.350 | 0.525 |
| He | 0.600 | 0.250 | 0.300 | 0.400 | 0.250 | 0.300 | 0.600 | 0.750 | 0.650 | 0.650 | 0.475 | |
| 平均值 Mean | Ho | 0.507 | 0.400 | 0.400 | 0.436 | 0.471 | 0.514 | 0.321 | 0.371 | 0.321 | 0.528 | 0.427 |
| He | 0.493 | 0.600 | 0.600 | 0.564 | 0.429 | 0.550 | 0.679 | 0.628 | 0.678 | 0.471 | 0.569 | |
| 位点 Locus | Fis | Fit | Fst | Nm |
|---|---|---|---|---|
| AAP-1 | -0.462 2 | -0.422 3 | 0.027 3 | 8.920 6 |
| SHDH | -0.365 0 | -0.245 9 | 0.087 2 | 2.615 7 |
| GOT-2 | -0.512 6 | -0.500 6 | 0.007 9 | 31.340 8 |
| PGI-4 | -0.506 9 | -0.486 8 | 0.013 3 | 18.552 9 |
| 6PGD-1 | -0.397 4 | -0.250 0 | 0.105 5 | 2.120 4 |
| MNR-3 | -0.234 9 | -0.223 0 | 0.009 6 | 25.692 8 |
| PGM-2 | -0.221 1 | -0.167 7 | 0.043 7 | 5.467 3 |
| Mean | -0.397 7 | -0.340 0 | 0.041 3 | 5.808 9 |
表6 麻疯树天然种群F统计量及基因流
Table 6 Estimated F-statistics and gene flow in natural population of Jatropha curcas
| 位点 Locus | Fis | Fit | Fst | Nm |
|---|---|---|---|---|
| AAP-1 | -0.462 2 | -0.422 3 | 0.027 3 | 8.920 6 |
| SHDH | -0.365 0 | -0.245 9 | 0.087 2 | 2.615 7 |
| GOT-2 | -0.512 6 | -0.500 6 | 0.007 9 | 31.340 8 |
| PGI-4 | -0.506 9 | -0.486 8 | 0.013 3 | 18.552 9 |
| 6PGD-1 | -0.397 4 | -0.250 0 | 0.105 5 | 2.120 4 |
| MNR-3 | -0.234 9 | -0.223 0 | 0.009 6 | 25.692 8 |
| PGM-2 | -0.221 1 | -0.167 7 | 0.043 7 | 5.467 3 |
| Mean | -0.397 7 | -0.340 0 | 0.041 3 | 5.808 9 |
| 种群代号 Population code | YL | PJ | HPZ | LB | FY | SB | XSBN | YNPE | HL | LS |
|---|---|---|---|---|---|---|---|---|---|---|
| YL | **** | 0.943 6 | 0.956 1 | 0.973 9 | 0.955 2 | 0.921 7 | 0.943 4 | 0.977 7 | 0.961 7 | 0.990 9 |
| PJ | 0.058 1 | **** | 0.995 3 | 0.958 1 | 0.961 7 | 0.941 2 | 0.975 3 | 0.964 5 | 0.959 1 | 0.945 0 |
| HPZ | 0.044 9 | 0.004 7 | **** | 0.964 1 | 0.969 9 | 0.941 8 | 0.972 1 | 0.967 6 | 0.962 8 | 0.962 4 |
| LB | 0.026 4 | 0.042 8 | 0.036 5 | **** | 0.979 3 | 0.983 3 | 0.980 7 | 0.990 2 | 0.980 0 | 0.974 3 |
| FY | 0.045 8 | 0.039 0 | 0.030 5 | 0.020 9 | **** | 0.969 4 | 0.979 3 | 0.968 1 | 0.977 6 | 0.966 8 |
| SB | 0.081 5 | 0.060 6 | 0.059 9 | 0.016 9 | 0.031 1 | **** | 0.981 0 | 0.965 3 | 0.970 7 | 0.928 4 |
| XSBN | 0.058 3 | 0.025 0 | 0.028 3 | 0.019 5 | 0.020 9 | 0.019 2 | **** | 0.979 3 | 0.989 5 | 0.953 8 |
| YNPE | 0.022 6 | 0.036 1 | 0.033 0 | 0.009 8 | 0.032 4 | 0.035 3 | 0.021 0 | **** | 0.987 2 | 0.975 6 |
| HL | 0.039 0 | 0.041 8 | 0.037 9 | 0.020 2 | 0.022 7 | 0.029 7 | 0.010 5 | 0.012 9 | **** | 0.968 7 |
| LS | 0.009 1 | 0.056 6 | 0.038 3 | 0.026 0 | 0.033 7 | 0.074 3 | 0.047 3 | 0.024 8 | 0.031 8 | **** |
表7 麻疯树种群间的遗传距离D (对角线下方)及遗传一致度I (对角线上方) (Nei, 1978)
Table 7 Genetic distances (below diagonal) and genetic identities (above diagonal) among populations of Jatropha curcas (Nei, 1978)
| 种群代号 Population code | YL | PJ | HPZ | LB | FY | SB | XSBN | YNPE | HL | LS |
|---|---|---|---|---|---|---|---|---|---|---|
| YL | **** | 0.943 6 | 0.956 1 | 0.973 9 | 0.955 2 | 0.921 7 | 0.943 4 | 0.977 7 | 0.961 7 | 0.990 9 |
| PJ | 0.058 1 | **** | 0.995 3 | 0.958 1 | 0.961 7 | 0.941 2 | 0.975 3 | 0.964 5 | 0.959 1 | 0.945 0 |
| HPZ | 0.044 9 | 0.004 7 | **** | 0.964 1 | 0.969 9 | 0.941 8 | 0.972 1 | 0.967 6 | 0.962 8 | 0.962 4 |
| LB | 0.026 4 | 0.042 8 | 0.036 5 | **** | 0.979 3 | 0.983 3 | 0.980 7 | 0.990 2 | 0.980 0 | 0.974 3 |
| FY | 0.045 8 | 0.039 0 | 0.030 5 | 0.020 9 | **** | 0.969 4 | 0.979 3 | 0.968 1 | 0.977 6 | 0.966 8 |
| SB | 0.081 5 | 0.060 6 | 0.059 9 | 0.016 9 | 0.031 1 | **** | 0.981 0 | 0.965 3 | 0.970 7 | 0.928 4 |
| XSBN | 0.058 3 | 0.025 0 | 0.028 3 | 0.019 5 | 0.020 9 | 0.019 2 | **** | 0.979 3 | 0.989 5 | 0.953 8 |
| YNPE | 0.022 6 | 0.036 1 | 0.033 0 | 0.009 8 | 0.032 4 | 0.035 3 | 0.021 0 | **** | 0.987 2 | 0.975 6 |
| HL | 0.039 0 | 0.041 8 | 0.037 9 | 0.020 2 | 0.022 7 | 0.029 7 | 0.010 5 | 0.012 9 | **** | 0.968 7 |
| LS | 0.009 1 | 0.056 6 | 0.038 3 | 0.026 0 | 0.033 7 | 0.074 3 | 0.047 3 | 0.024 8 | 0.031 8 | **** |
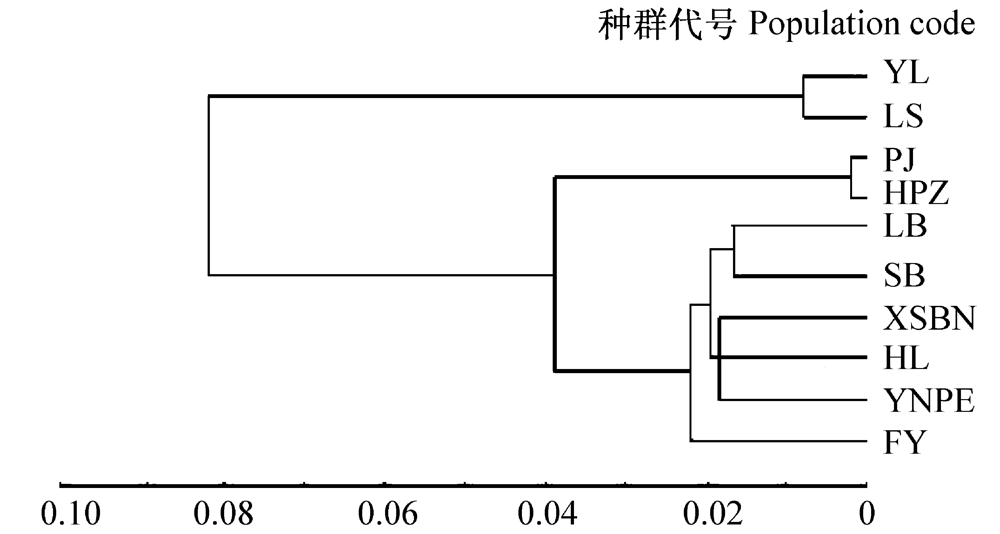
图1 麻疯树种群的Nei’s遗传距离UPGMA聚类图。种群代号同表1。
Fig. 1 UPGMA dendrogram of Jatropha curcas populations based on Nei’s genetic distance. Population codes see Table 1.
| [1] |
Basha SD, Francis G, Makkar HPS, Becker K, Sujatha M (2009). A comparative study of biochemical traits and molecular markers for assessment of genetic relationships between Jatropha curcas L. germplasm from different countries. Plant Science, 176, 812-823.
DOI URL |
| [2] | Chen SY (陈少瑜), Wu LY (吴丽圆), Li JW (李江文), Xiang W (项伟), Zhou Y (周云) (2001). Study on genetic diversity of natural population of Taxus yunanensis. Scientia Silvae Sinicae (林业科学), 37(5), 41-48. (in Chinese with English abstract) |
| [3] | Ge S (葛颂) (1989). Quantitative analysis of tree population variation by allozyme and analysis method. Journal of Southwest Forestry College (西南林学院学报), 9(1), 84-91. (in Chinese with English abstract) |
| [4] | Gu YJ (辜云杰), He CJ (何朝军), Zhang J (张俊), He SB (何绍彬), Zhang W (张炜), Luo JX (罗建勋) (2009). Study on phenotypic diversity of Jatropha curas seeds from natural population in Liangshan Yi Autonomous Prefecture. Journal of Southwest Forestry College (西南林学院学报), 29(6), 11-14. (in Chinese with English abstract) |
| [5] | Hamrick JL, Godt MJW (1989). Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS eds. Plant Population Genetics, Breeding and Germplasm Resources. Sinauer, Sunderland, Mass. 43-63. |
| [6] |
Hamrick JL, Godt MJW, Sherman-Bmyles SL (1992). Factors influencing levels of genetic diversity in woody plant species. New Forests, 6, 95-124.
DOI URL |
| [7] | He W (何玮), Guo L (郭亮), Wang L (王岚), Yang W (杨威), Tang L (唐琳), Chen F (陈放) (2007). ISSR analysis of genetic diversity of Jatropha curcas L. Chinese Journal of Applied and Environmental Biology (应用与环境生物学报), 13, 466-470. (in Chinese with English abstract) |
| [8] | Kaushik N, Kumar K, Kumar S, Kaushik N, Roy S (2007). Genetic variability and divergence studies in seed traits and oil content of Jatropha (Jatropha curcas L.) accessions. Biomass & Bioenergy, 31, 497-502. |
| [9] | Luo JX (罗建勋), Gu YJ (辜云杰), Tang P (唐平), He XF (和献峰), Peng JY (彭建勇), Zhang W (张炜) (2009). Jatropha curcas populations in west Panzhihua area. Journal of Southwest Forestry College (西南林学院学报), 29(3), 1-4. (in Chinese with English abstract) |
| [10] | Luo JX (罗建勋), He XF (和献峰), Gu YJ (辜云杰), Tang P (唐平), Cao XJ (曹小军), Yang PJ (杨培金) (2008). Study on phenotypic diversity of natural populations of Jatropha curcas in Panzhihua area, Sichuan Province. Journal of Southwest Forestry College (西南林学院学报), 28(6), 31-35. (in Chinese with English abstract) |
| [11] |
Nei M (1972). Genetic distance between populations. The American Naturalist, 6, 283-293.
DOI URL |
| [12] |
Nei M (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89, 583-590.
URL PMID |
| [13] | Ou WJ (欧文军), Wang WQ (王文泉), Li KM (李开绵) (2009). Molecular genetic diversity analysis of 120 accessions of Jatropha curcas L. germplasm. Chinese Journal of Tropical Crops (热带作物学报), 30, 284-292. (in Chinese with English abstract) |
| [14] |
Soltis DE, Haufler CH, Darow DC, Gastony GJ (1983). Starch gel electrophoresis of ferns: a compilation of grinding buffers, gel and electrode buffers, and staining schedules. American Fern Journal, 73, 9-27.
DOI URL |
| [15] |
Swofford DL, Selander RB (1989). BIOSIS-1, a computer program for the analysis of allelic variation in population genetics and biochemical systematics. Journal of Heredity, 72, 281-283.
DOI URL |
| [16] |
Tatikonda L, Wani SP, Kannan S, Beerelli N, Sreedevi TK, Hoisington DA, Devi P, Varshney RK (2009). AFLP- based molecular characterization of an elite germplasm collection of Jatropha curcas L., a biofuel plant. Plant Science, 176, 505-513.
DOI URL PMID |
| [17] | Wang ZR (王中仁) 1996. Plant Allozyme Analysis 植物等位酶分析. Science Press, Beijing. (in Chinese) |
| [18] | Wendel JF, Weeden NF (1989). Visualization and interpretation of plant isozymes. In: Soltis DE, Soltis PS eds. Isozymes in Plant Biology. Dioscordes Press, Portland. 5-44. |
| [19] | Xiang ZY (向振勇), Song SQ (宋松泉), Wang GJ (王桂娟), Chen MS (陈茂盛), Yang CY (杨成源), Long CL (龙春林) (2007). Genetic diversity of Jatropha curcas (Euphorbiaceae) collected from southern Yunnan, detected by inter-simple sequence repeat (ISSR). Acta Botanica Yunnanic (云南植物研究), 29, 619-624. (in Chinese with English abstract) |
| [20] | Yeh FC, Yang RC, Boyle T (1997). POPGENE, Version 1.32 Edition. Software Microsoft Window-Based Freeware for Population Genetic Analysis. University of Alberta, Edmonton, Alta. |
| [1] | 陈天翌, 娄安如. 青藏高原东侧白桦种群的遗传多样性与遗传结构[J]. 植物生态学报, 2022, 46(5): 561-568. |
| [2] | 张新新, 王茜, 胡颖, 周玮, 陈晓阳, 胡新生. 植物边缘种群遗传多样性研究进展[J]. 植物生态学报, 2019, 43(5): 383-395. |
| [3] | 张俪文, 韩广轩. 植物遗传多样性与生态系统功能关系的研究进展[J]. 植物生态学报, 2018, 42(10): 977-989. |
| [4] | 王锦楠, 陈进福, 陈武生, 周新洋, 许东, 李际红, 亓晓. 柴达木地区野生黑果枸杞种群遗传多样性的AFLP分析[J]. 植物生态学报, 2015, 39(10): 1003-1011. |
| [5] | 刘军, 姜景民, 邹军, 徐金良, 沈汉, 刁松峰. 中国特有濒危树种毛红椿核心和边缘居群的遗传多样性[J]. 植物生态学报, 2013, 37(1): 52-60. |
| [6] | 魏源, 王世杰, 刘秀明, 黄天志. 不同喀斯特小生境中土壤丛枝菌根真菌的遗传多样性[J]. 植物生态学报, 2011, 35(10): 1083-1090. |
| [7] | 张云红, 侯艳, 娄安如. 华北地区小丛红景天种群的AFLP遗传多样性[J]. 植物生态学报, 2010, 34(9): 1084-1094. |
| [8] | 刘伟, 王曦, 干友民, 黄林凯, 谢文刚, 苗佳敏. 高山嵩草种群在放牧干扰下遗传多样性的变化[J]. 植物生态学报, 2009, 33(5): 966-973. |
| [9] | 陈良华, 胡庭兴, 张帆, 李国和. 用AFLP技术分析四川核桃资源的遗传多样性[J]. 植物生态学报, 2008, 32(6): 1362-1372. |
| [10] | 周会平, 陈进, 张寿洲. 具混合繁殖策略的草本植物异果舞花姜的居群遗传结构[J]. 植物生态学报, 2008, 32(4): 751-759. |
| [11] | 严茂粉, 李向华, 王克晶. 北京地区野生大豆种群SSR标记的遗传多样性评价[J]. 植物生态学报, 2008, 32(4): 938-950. |
| [12] | 刘亚令, 李作洲, 姜正旺, 刘义飞, 黄宏文. 中华猕猴桃和美味猕猴桃自然居群遗传结构及其种间杂交渐渗[J]. 植物生态学报, 2008, 32(3): 704-718. |
| [13] | 魏宇昆, 高玉葆. 禾草内生真菌的遗传多样性及其共生关系[J]. 植物生态学报, 2008, 32(2): 512-520. |
| [14] | 杨明博, 杨劼, 杨九艳, 梁娜, 清华. 鄂尔多斯高原不同生境条件下中间锦鸡儿植物叶片表皮特征及遗传多样性变化分析[J]. 植物生态学报, 2007, 31(6): 1181-1189. |
| [15] | 穆立蔷, 刘赢男. 不同地理分布区紫椴种群的遗传多样性变化[J]. 植物生态学报, 2007, 31(6): 1190-1198. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
Copyright © 2022 版权所有 《植物生态学报》编辑部
地址: 北京香山南辛村20号, 邮编: 100093
Tel.: 010-62836134, 62836138; Fax: 010-82599431; E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
备案号: 京ICP备16067583号-19