

植物生态学报 ›› 2008, Vol. 32 ›› Issue (3): 704-718.DOI: 10.3773/j.issn.1005-264x.2008.03.021
刘亚令1,2( ), 李作洲2, 姜正旺2, 刘义飞2, 黄宏文1,2,*(
), 李作洲2, 姜正旺2, 刘义飞2, 黄宏文1,2,*( )
)
收稿日期:2006-09-12
接受日期:2006-12-11
出版日期:2008-09-12
发布日期:2008-05-30
通讯作者:
黄宏文
作者简介:*E-mail:hongwen@public.wh.hb.cn基金资助:
LIU Ya-Ling1,2( ), LI Zuo-Zhou2, JIANG Zheng-Wang2, LIU Yi-Fei2, HUANG Hong-Wen1,2,*(
), LI Zuo-Zhou2, JIANG Zheng-Wang2, LIU Yi-Fei2, HUANG Hong-Wen1,2,*( )
)
Received:2006-09-12
Accepted:2006-12-11
Online:2008-09-12
Published:2008-05-30
Contact:
HUANG Hong-Wen
摘要:
利用9对SSR引物对中华猕猴桃(Actinidia chinensis)和美味猕猴桃(A. deliciosa)两近缘种的5个同域分布复合体和各自1个非同域分布居群进行了遗传多样性、居群遗传结构的分析以及种间杂交渐渗的探讨。结果表明:1)两物种共有等位基因比例高达81.13%,物种特有等位基因较少(中华猕猴桃:13.27%,美味猕猴桃:5.61%),但共享等位基因表型频率在两近缘种间存在差异,而且与各同域复合体中两物种样本的交错程度或间距存在关联;2)两种猕猴桃均具有极高遗传多样性,美味猕猴桃的遗传多样性(Ho=0.749,PIC=0.818)都略高于中华猕猴桃(Ho=0.686,PIC=0.799);3)两猕猴桃物种均具有较低的Nei's居群遗传分化度,但AMOVA分析结果揭示种内异域居群间(FST=0.091 5)和同域复合体种间(FST=0.111 5)均存在一定程度的遗传分化;中华猕猴桃居群遗传分化(GST=0.086; FST=0.212 1)高于美味猕猴桃(GST=0.080; FST=0.142 0);4)同域分布复合体两物种间的遗传分化(GST=0.020)低于物种内异域居群间的遗传分化(中华猕猴桃:GST=0.086; 美味猕猴桃:GST=0.080),同域复合体物种间的基因流(Nm=7.89-29.75)远远高于同种异域居群间(中华猕猴桃:Nm =2.663; 美味猕猴桃:Nm =2.880);5)居群UPGMA聚类揭示在同一地域的居群优先聚类,个体聚类结果显示多数个体聚在各自居群组内,但各地理居群并不按地理距离的远近聚类,这与Mantel相关性检测所揭示的居群间遗传距离与地理距离没有显著性相关的结果一致。进一步分析表明两种猕猴桃的遗传多样性和居群遗传结构不仅受其广域分布、远交、晚期分化等生活史特性的影响,同时还与猕猴桃的染色体基数高(x=29)、倍性复杂和种间杂交等因素密切相关,其中两种猕猴桃的共享祖先多态性和同域分布种间杂交基因渗透对两猕猴桃的居群遗传结构产生了重要影响。
刘亚令, 李作洲, 姜正旺, 刘义飞, 黄宏文. 中华猕猴桃和美味猕猴桃自然居群遗传结构及其种间杂交渐渗. 植物生态学报, 2008, 32(3): 704-718. DOI: 10.3773/j.issn.1005-264x.2008.03.021
LIU Ya-Ling, LI Zuo-Zhou, JIANG Zheng-Wang, LIU Yi-Fei, HUANG Hong-Wen. GENETIC STRUCTURE AND HYBRIDIZATION INTROGRESSION IN NATURAL POPULATIONS OF TWO CLOSELY RELATED ACTINIDIA SPECIES, A. CHINENSIS AND A. DELICIOSA. Chinese Journal of Plant Ecology, 2008, 32(3): 704-718. DOI: 10.3773/j.issn.1005-264x.2008.03.021
| 位点 Loci | 等位基因数 No. of alleles | 共有等位基因比率 Percentage of common alleles (%) | 共有等位基因数 Common alleles in both species | 特有等位基因数 Species specific alleles | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 中华猕猴桃 Actinidia chinensis | 美味猕猴桃 A. deliciosa | ||||||||||||||||||||||||||||||||||
| UDK96-001 | 36 | 86.1 | 31 | 3 | 2 | ||||||||||||||||||||||||||||||
| UDK96-009 | 21 | 66.7 | 14 | 5 | 2 | ||||||||||||||||||||||||||||||
| UDK96-013 | 17 | 58.8 | 10 | 6 | 1 | ||||||||||||||||||||||||||||||
| UDK96-019 | 27 | 92.6 | 25 | 1 | 1 | ||||||||||||||||||||||||||||||
| UDK96-028 | 17 | 64.7 | 11 | 5 | 1 | ||||||||||||||||||||||||||||||
| UDK96-039 | 18 | 83.3 | 15 | 0 | 3 | ||||||||||||||||||||||||||||||
| UDK97-404 | 19 | 89.5 | 17 | 1 | 1 | ||||||||||||||||||||||||||||||
| UDK97-408 | 21 | 90.5 | 19 | 2 | 0 | ||||||||||||||||||||||||||||||
| UDK97-420 | 20 | 85.0 | 17 | 3 | 0 | ||||||||||||||||||||||||||||||
| 共计Total | 196 | 81.1 | 159 | 26 | 11 | ||||||||||||||||||||||||||||||
| 平均Mean | 21.78 | 79.7 | 17.67 | 2.89 | 1.22 | ||||||||||||||||||||||||||||||
表1 中华猕猴桃和美味猕猴桃的共有及特有等位基因数
Table 1 The number of common and specific alleles of Actinidia chinensis and A. deliciosa
| 位点 Loci | 等位基因数 No. of alleles | 共有等位基因比率 Percentage of common alleles (%) | 共有等位基因数 Common alleles in both species | 特有等位基因数 Species specific alleles | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 中华猕猴桃 Actinidia chinensis | 美味猕猴桃 A. deliciosa | ||||||||||||||||||||||||||||||||||
| UDK96-001 | 36 | 86.1 | 31 | 3 | 2 | ||||||||||||||||||||||||||||||
| UDK96-009 | 21 | 66.7 | 14 | 5 | 2 | ||||||||||||||||||||||||||||||
| UDK96-013 | 17 | 58.8 | 10 | 6 | 1 | ||||||||||||||||||||||||||||||
| UDK96-019 | 27 | 92.6 | 25 | 1 | 1 | ||||||||||||||||||||||||||||||
| UDK96-028 | 17 | 64.7 | 11 | 5 | 1 | ||||||||||||||||||||||||||||||
| UDK96-039 | 18 | 83.3 | 15 | 0 | 3 | ||||||||||||||||||||||||||||||
| UDK97-404 | 19 | 89.5 | 17 | 1 | 1 | ||||||||||||||||||||||||||||||
| UDK97-408 | 21 | 90.5 | 19 | 2 | 0 | ||||||||||||||||||||||||||||||
| UDK97-420 | 20 | 85.0 | 17 | 3 | 0 | ||||||||||||||||||||||||||||||
| 共计Total | 196 | 81.1 | 159 | 26 | 11 | ||||||||||||||||||||||||||||||
| 平均Mean | 21.78 | 79.7 | 17.67 | 2.89 | 1.22 | ||||||||||||||||||||||||||||||
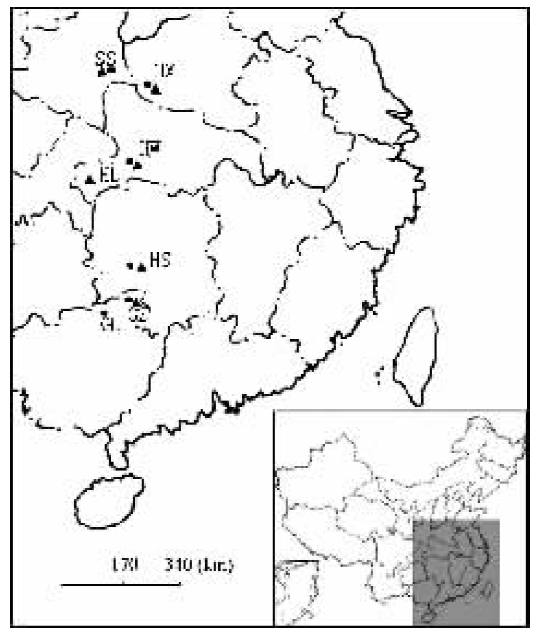
图1 中华猕猴桃居群和美味猕猴桃居群的采样图 SS:陕西省商南县Shangnan County of Shaanxi Province HX:河南省西峡县Xixia County of Henan Province HW:湖北省五峰县Wufeng County of Hubei Province HL:湖北省利川县Lichuan County of Hubei Province HS:湖南省绥宁县Suining County of Hunan Province GZ:广西省资源县Ziyuan County of Guangxi Province GL:广西省龙胜县Longsheng County of Guangxi Province ■:中华猕猴桃Actinidia chinensis ▲:美味猕猴桃A. deliciosa
Fig.1 Map of the sampled populations of Actinidia chinensis and A. deliciosa
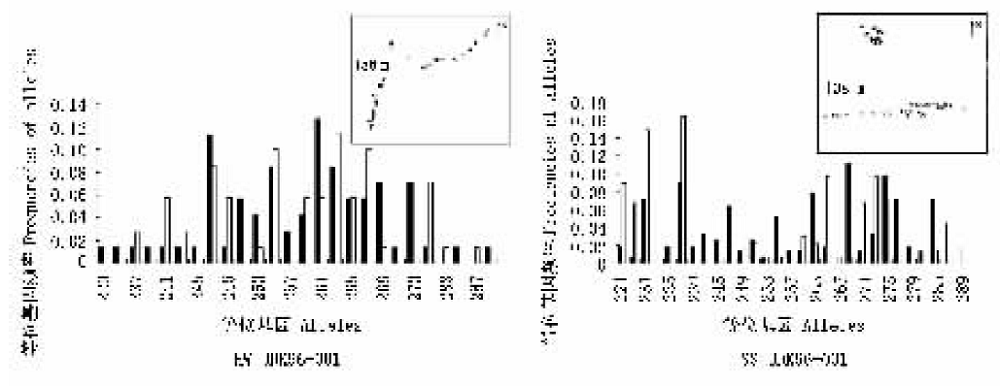
图2 中华猕猴桃(白色)和美味猕猴桃(黑色)物种的SSR片段频率在位点UDK96-001上的分布图及两物种在湖北五峰(HW)和陕西商南(SS)的个体分布图
Fig.2 Histograms illustration the frequency distributions of microsatellite length fragments in Actinidia chinensis (white bars) and A. deliciosa (black bars) for locus UDK96-001 and the natural distribution of sampled individuals of two Actinidia species in Wufeng, Hubei (HW) and Shangnan, Shaanxi (SS)
| 居群 Population | 中华猕猴桃Actinidia chinensis | 美味猕猴桃A. deliciosa | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | A | P | Ho | PIC | N | A | P | Ho | PIC | |
| 湖北五峰Wufeng Hubei (HW) | 20 | 12.11 | 100 | 0.922 | 0.834 | 31 | 12.78 | 100 | 0.703 | 0.832 |
| 广西资源Ziyuan Guangxi (GZ) | 36 | 11.56 | 100 | 0.423 | 0.757 | 11 | 7.33 | 100 | 0.545 | 0.790 |
| 河南西峡Xixia Henan (HX) | 52 | 14.44 | 100 | 0.891 | 0.865 | 39 | 13.33 | 100 | 0.826 | 0.841 |
| 陕西商南Shangnan Shaanxi (SS) | 47 | 12.22 | 100 | 0.768 | 0.758 | 49 | 13.33 | 100 | 0.802 | 0.820 |
| 湖南绥宁Suining Hunan (HS) | 35 | 12.89 | 100 | 0.724 | 0.823 | 35 | 12.89 | 100 | 0.781 | 0.781 |
| 广西龙胜Longsheng Guangxi (GL) | 20 | 6.67 | 100 | 0.389 | 0.765 | |||||
| 湖北利川Lichuan Hubei (HL) | 57 | 15.11 | 100 | 0.836 | 0.846 | |||||
| 居群平均Mean at population level | 35 | 11.65 | 100 | 0.686 | 0.800 | 37 | 12.46 | 100 | 0.749 | 0.818 |
| 标准误差 Standard error | 2.40 | 0.209 | 0.042 | 2.42 | 0.101 | 0.025 | ||||
| 物种水平At species level | 210 | 20.56 | 100 | 0.711 | 0.874 | 222 | 18.89 | 100 | 0.782 | 0.889 |
表2 两种猕猴桃的居群遗传多样性
Table 2 Population genetic diversity of Actinidia chinensis and A. deliciosa
| 居群 Population | 中华猕猴桃Actinidia chinensis | 美味猕猴桃A. deliciosa | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | A | P | Ho | PIC | N | A | P | Ho | PIC | |
| 湖北五峰Wufeng Hubei (HW) | 20 | 12.11 | 100 | 0.922 | 0.834 | 31 | 12.78 | 100 | 0.703 | 0.832 |
| 广西资源Ziyuan Guangxi (GZ) | 36 | 11.56 | 100 | 0.423 | 0.757 | 11 | 7.33 | 100 | 0.545 | 0.790 |
| 河南西峡Xixia Henan (HX) | 52 | 14.44 | 100 | 0.891 | 0.865 | 39 | 13.33 | 100 | 0.826 | 0.841 |
| 陕西商南Shangnan Shaanxi (SS) | 47 | 12.22 | 100 | 0.768 | 0.758 | 49 | 13.33 | 100 | 0.802 | 0.820 |
| 湖南绥宁Suining Hunan (HS) | 35 | 12.89 | 100 | 0.724 | 0.823 | 35 | 12.89 | 100 | 0.781 | 0.781 |
| 广西龙胜Longsheng Guangxi (GL) | 20 | 6.67 | 100 | 0.389 | 0.765 | |||||
| 湖北利川Lichuan Hubei (HL) | 57 | 15.11 | 100 | 0.836 | 0.846 | |||||
| 居群平均Mean at population level | 35 | 11.65 | 100 | 0.686 | 0.800 | 37 | 12.46 | 100 | 0.749 | 0.818 |
| 标准误差 Standard error | 2.40 | 0.209 | 0.042 | 2.42 | 0.101 | 0.025 | ||||
| 物种水平At species level | 210 | 20.56 | 100 | 0.711 | 0.874 | 222 | 18.89 | 100 | 0.782 | 0.889 |
| 位点 Loci | 中华猕猴桃Actinidia chinensis | 美味猕猴桃A. deliciosa | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HT | HS | DST | GST | Nm | HT | HS | DST | GST | Nm | |
| UDK96-001 | 0.940 | 0.887 | 0.053 | 0.056 | 4.184 | 0.956 | 0.905 | 0.051 | 0.053 | 4.436 |
| UDK96-009 | 0.857 | 0.721 | 0.136 | 0.159 | 1.325 | 0.962 | 0.701 | 0.261 | 0.271 | 0.671 |
| UDK96-013 | 0.879 | 0.788 | 0.091 | 0.104 | 2.165 | 0.879 | 0.794 | 0.085 | 0.097 | 2.354 |
| UDK96-019 | 0.944 | 0.908 | 0.036 | 0.038 | 6.306 | 0.929 | 0.906 | 0.023 | 0.025 | 9.848 |
| UDK96-028 | 0.700 | 0.542 | 0.158 | 0.226 | 0.858 | 0.698 | 0.597 | 0.101 | 0.145 | 1.478 |
| UDK96-039 | 0.841 | 0.768 | 0.073 | 0.087 | 2.630 | 0.870 | 0.830 | 0.040 | 0.046 | 5.188 |
| UDK97-404 | 0.898 | 0.858 | 0.040 | 0.045 | 5.363 | 0.893 | 0.879 | 0.014 | 0.016 | 15.696 |
| UDK97-408 | 0.895 | 0.845 | 0.050 | 0.056 | 4.225 | 0.895 | 0.869 | 0.026 | 0.029 | 8.356 |
| UDK97-420 | 0.910 | 0.871 | 0.039 | 0.043 | 5.583 | 0.916 | 0.882 | 0.034 | 0.037 | 6.485 |
| 平均Mean (标准误差SE) | 0.874 (0.066) | 0.799 (0.102) | 0.075 (0.040) | 0.086 | 2.663 | 0.889 (0.070) | 0.818 (0.094) | 0.071 (0.069) | 0.080 | 2.880 |
表3 中华猕猴桃和美味猕猴桃种内居群遗传分化
Table 3 The genetic differentiation of Actinidia chinensis and A. deliciosa
| 位点 Loci | 中华猕猴桃Actinidia chinensis | 美味猕猴桃A. deliciosa | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HT | HS | DST | GST | Nm | HT | HS | DST | GST | Nm | |
| UDK96-001 | 0.940 | 0.887 | 0.053 | 0.056 | 4.184 | 0.956 | 0.905 | 0.051 | 0.053 | 4.436 |
| UDK96-009 | 0.857 | 0.721 | 0.136 | 0.159 | 1.325 | 0.962 | 0.701 | 0.261 | 0.271 | 0.671 |
| UDK96-013 | 0.879 | 0.788 | 0.091 | 0.104 | 2.165 | 0.879 | 0.794 | 0.085 | 0.097 | 2.354 |
| UDK96-019 | 0.944 | 0.908 | 0.036 | 0.038 | 6.306 | 0.929 | 0.906 | 0.023 | 0.025 | 9.848 |
| UDK96-028 | 0.700 | 0.542 | 0.158 | 0.226 | 0.858 | 0.698 | 0.597 | 0.101 | 0.145 | 1.478 |
| UDK96-039 | 0.841 | 0.768 | 0.073 | 0.087 | 2.630 | 0.870 | 0.830 | 0.040 | 0.046 | 5.188 |
| UDK97-404 | 0.898 | 0.858 | 0.040 | 0.045 | 5.363 | 0.893 | 0.879 | 0.014 | 0.016 | 15.696 |
| UDK97-408 | 0.895 | 0.845 | 0.050 | 0.056 | 4.225 | 0.895 | 0.869 | 0.026 | 0.029 | 8.356 |
| UDK97-420 | 0.910 | 0.871 | 0.039 | 0.043 | 5.583 | 0.916 | 0.882 | 0.034 | 0.037 | 6.485 |
| 平均Mean (标准误差SE) | 0.874 (0.066) | 0.799 (0.102) | 0.075 (0.040) | 0.086 | 2.663 | 0.889 (0.070) | 0.818 (0.094) | 0.071 (0.069) | 0.080 | 2.880 |
| 变异来源 Source of variation | 自由度 df | 离差平方和 Sum of squared deviations (SSD) | 方差分量 Variance component | 方差分量比率 Percentage of variation (%) | 固定指数 Fixation index* | p |
|---|---|---|---|---|---|---|
| 种间 Among species | 1 | 168.33 | 0.18 | 1.00 | FCT=0.01 | 0.229 |
| 种内居群间Among population within species | 10 | 1 150.49 | 2.90 | 17.31 | FSC=0.18 | <0.001 |
| 居群内 Within populations | 420 | 5 738.23 | 13.67 | 81.68 | FST=0.18 | <0.001 |
| 总和 Total | 431 | 7 057.05 | 16.73 | 100.00 | - | - |
| 中华猕猴桃居群Actinidia chinensis populations group | ||||||
| 居群间 Among populations | 5 | 636.06 | 3.37 | 21.61 | FST=0.21 | <0.001 |
| 居群内 Within population | 204 | 2 491.30 | 12.21 | 78.39 | - | <0.001 |
| 总和 Total | 209 | 3 127.36 | 15.58 | 100.000 | - | - |
| 美味猕猴桃居群 A. deliciosa populations group | ||||||
| 居群间 Among populations | 5 | 521.55 | 2.49 | 14.20 | FST=0.14 | <0.001 |
| 居群内 Within population | 216 | 3 248.67 | 15.04 | 85.80 | - | <0.001 |
| 总和 Total | 221 | 3 770.22 | 17.53 | 100.00 | - | - |
表4 中华猕猴桃和美味猕猴桃居群的AMOVA分析
Table 4 AMOVA analysis of Actinidia chinensis and A. deliciosa populations
| 变异来源 Source of variation | 自由度 df | 离差平方和 Sum of squared deviations (SSD) | 方差分量 Variance component | 方差分量比率 Percentage of variation (%) | 固定指数 Fixation index* | p |
|---|---|---|---|---|---|---|
| 种间 Among species | 1 | 168.33 | 0.18 | 1.00 | FCT=0.01 | 0.229 |
| 种内居群间Among population within species | 10 | 1 150.49 | 2.90 | 17.31 | FSC=0.18 | <0.001 |
| 居群内 Within populations | 420 | 5 738.23 | 13.67 | 81.68 | FST=0.18 | <0.001 |
| 总和 Total | 431 | 7 057.05 | 16.73 | 100.00 | - | - |
| 中华猕猴桃居群Actinidia chinensis populations group | ||||||
| 居群间 Among populations | 5 | 636.06 | 3.37 | 21.61 | FST=0.21 | <0.001 |
| 居群内 Within population | 204 | 2 491.30 | 12.21 | 78.39 | - | <0.001 |
| 总和 Total | 209 | 3 127.36 | 15.58 | 100.000 | - | - |
| 美味猕猴桃居群 A. deliciosa populations group | ||||||
| 居群间 Among populations | 5 | 521.55 | 2.49 | 14.20 | FST=0.14 | <0.001 |
| 居群内 Within population | 216 | 3 248.67 | 15.04 | 85.80 | - | <0.001 |
| 总和 Total | 221 | 3 770.22 | 17.53 | 100.00 | - | - |
| 居群 Population | HT | HS | DST | GST | Nm |
|---|---|---|---|---|---|
| HW | 0.840 | 0.833 | 0.007 | 0.008 | 29.750 |
| GZ | 0.787 | 0.773 | 0.014 | 0.017 | 13.804 |
| HX | 0.872 | 0.853 | 0.019 | 0.022 | 11.224 |
| SS | 0.814 | 0.789 | 0.025 | 0.031 | 7.890 |
| HS | 0.815 | 0.797 | 0.018 | 0.021 | 11.069 |
| 平均Mean(标准误差SE) | 0.826(0.026) | 0.809(0.027) | 0.017(0.005) | 0.020 | 14.747 |
表5 中华猕猴桃和美味猕猴桃同域分布复合体种间的遗传分化
Table 5 Genetic differentiation between Actinidia chinensis and A. deliciosa in five sympatric distributed species-complex populations
| 居群 Population | HT | HS | DST | GST | Nm |
|---|---|---|---|---|---|
| HW | 0.840 | 0.833 | 0.007 | 0.008 | 29.750 |
| GZ | 0.787 | 0.773 | 0.014 | 0.017 | 13.804 |
| HX | 0.872 | 0.853 | 0.019 | 0.022 | 11.224 |
| SS | 0.814 | 0.789 | 0.025 | 0.031 | 7.890 |
| HS | 0.815 | 0.797 | 0.018 | 0.021 | 11.069 |
| 平均Mean(标准误差SE) | 0.826(0.026) | 0.809(0.027) | 0.017(0.005) | 0.020 | 14.747 |
| 变异来源 Source of variation | 自由度 df | 离差平方和 Sum of squared deviations (SSD) | 方差分量 Variance component | 方差分量比率 Percentage of variation (%) | 固定指数 Fixation index* | p |
|---|---|---|---|---|---|---|
| 中华/美味复合体地域间 Between regions | 4 | 734.66 | 1.54 | 9.16 | FCT=0.091 5 | <0.001 |
| 同地域复合体种间 Between species within sympatric population of the same region | 5 | 356.34 | 1.71 | 10.13 | FSC=0.111 5 | <0.001 |
| 居群内 Within population | 345 | 4 699.03 | 13.62 | 80.73 | FST=0.193 | <0.001 |
| 总和 Total | 354 | 5 790.02 | 16.87 | 100.00 | - | - |
表6 同域分布的中华猕猴桃和美味猕猴桃居群的AMOVA分析
Table 6 AMOVA analysis of sympatric distributed Actinidia chinensis and A. deliciosa populations
| 变异来源 Source of variation | 自由度 df | 离差平方和 Sum of squared deviations (SSD) | 方差分量 Variance component | 方差分量比率 Percentage of variation (%) | 固定指数 Fixation index* | p |
|---|---|---|---|---|---|---|
| 中华/美味复合体地域间 Between regions | 4 | 734.66 | 1.54 | 9.16 | FCT=0.091 5 | <0.001 |
| 同地域复合体种间 Between species within sympatric population of the same region | 5 | 356.34 | 1.71 | 10.13 | FSC=0.111 5 | <0.001 |
| 居群内 Within population | 345 | 4 699.03 | 13.62 | 80.73 | FST=0.193 | <0.001 |
| 总和 Total | 354 | 5 790.02 | 16.87 | 100.00 | - | - |
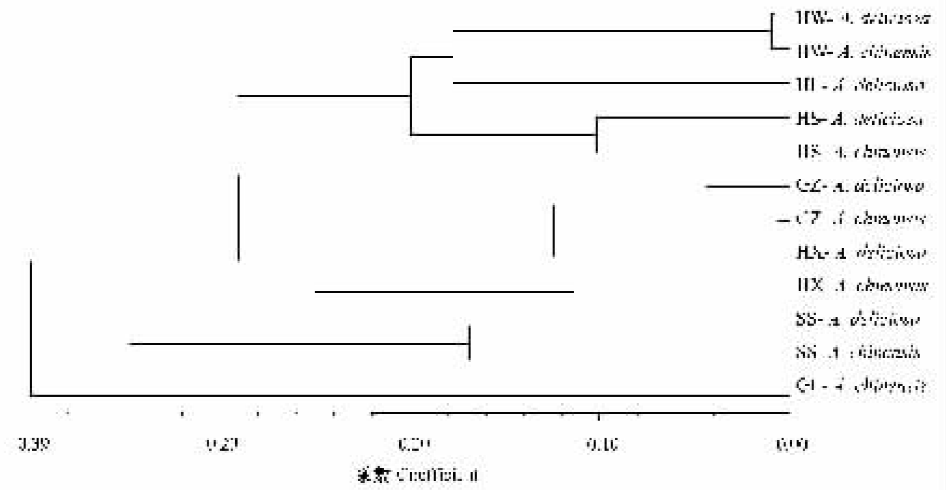
图3 中华猕猴桃和美味猕猴桃分布点的聚类分析树状图(UPGMA) HW、HL、HS、GZ、HX、SS、GL:见图1 See Fig. 1
Fig.3 Dendrogram of wild Actinidia chinensis and A. deliciosa based on SSR data (UPGMA)
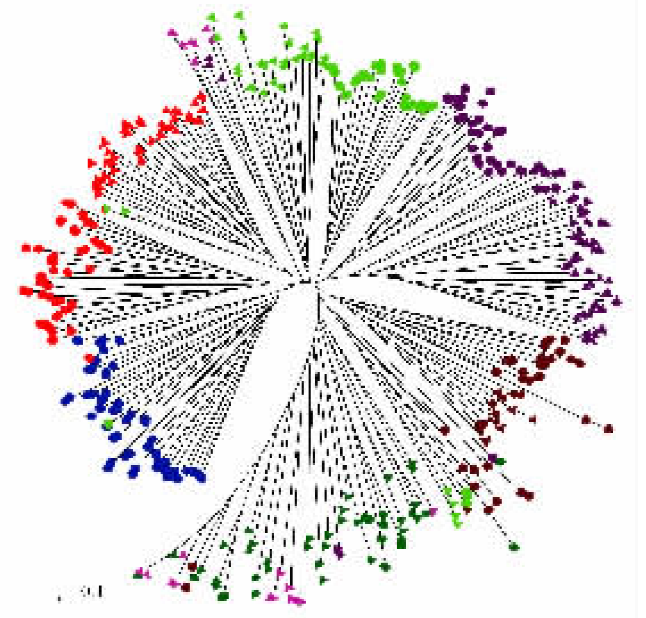
图4 基于9个SSR位点的中华猕猴桃(▲)和美味猕猴桃(●)的7个自然居群、432个植株的个体聚类种系树 居群的代表颜色为:HW(绛色);GZ(暗绿);HX(紫色);SS(红色);HS(绿色);HL(蓝色);GL(粉色) The colour represents different natural populations: HW (Crimson); GZ (Sap green); HX (Purple); SS (Red); HS (Green); HL (Blue); GL (Pink) HW、GZ、HX、SS、HS、HL、GL:见图1 See Fig. 1
Fig.4 Unrooted phylogeny tree of individual of Actinidia chinensis (▲)and A. deliciosa (●) based on 9 SSR loci (The tree contains 432 individuals from 7 natural populations)
| [1] | Aagaard SMD, Sastad SM, Greilhuber J, Moen A (2005). A secondary hybrid zone between diploid Dactylorhiza incarnate ssp. cruenta and allotetraploid D. lapponica (Orchidaceae). Heredity, 94,488-496. |
| [2] | Atkinson RG, Cipriani G, Whittaker DJ, Gardner RC (1997). The allopolyploid origin of kiwifruit, Actinidia deliciosa (Actinidiaceae). Plant Systematics and Evolution, 205,111-124. |
| [3] | Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Stuhl K (1987). Current Protocols in Molecular Biology. John Wiley & Sons, New York. |
| [4] | Barluenga M, Stølting KN, Salzburger W, Muschick M, Meyer A (2006). Sympatric speciation in Nicaraguan crater lake cichlid fish. Nature, 439,719-723. |
| [5] | Barton NH (2001). The role of hybridization in evolution. Molecular Ecology, 10,551-568. |
| [6] | Becher SA, Steinmetz K, Weising K, Boury S, Peltier D, Renou JP, Kahl G, Wolff K (2000). Microsatellites for cultivar identification in Pelargonium. Theoretical and Applied Genetics, 101,643-651. |
| [7] | Becker U, Reinhold T, Matthies D (2006). Effects of pollination distance on reproduction and offspring performance in Hypochoeris radicata: experiments with plants from three European regions . Biological Conservation, 132,109-118. |
| [8] | Bertin P, Grégoire D, Massart S, de Froidmont D (2001). Genetic diversity among European cultivated spelt revealed by microsatellites. Theoretical and Applied Genetics, 102,148-156. |
| [9] |
Bertin P, Grégoire D, Massart S, Froidmont D (2004). High level of genetic diversity among spelt germplasm revealed by microsatellite markers. Genome, 47,1043-1052.
URL PMID |
| [10] |
Bleeker W, Matthies A (2005). Hybrid zones between invasive Rorippa austriaca and native R. syivestris (Brassicaceae) in Germany: ploidy levels and patterns of fitness in the field . Heredity, 94,664-670.
URL PMID |
| [11] | Buteler MI, Jarret RL, Labonte DR (1999). Sequence characterization of microsatellites in diploid and polyploid Ipomoea. Theoretical and Applied Genetics, 99,123-132. |
| [12] | Callen DF, Thompson AD, Shen Y, Phillips HA, Richards RI, Mulley JC, Sutherland GR (1993). Incidence and origin of “null” alleles in the (AC) n microsatellite markers. American Journal of Human Genetics, 52,922-927. |
| [13] | Cantini C, Iezzoni AF, Lamboy WF, Boritzki M, Struss D (2001). DNA fingerprinting of tetraploid cherry germplasm using simple sequence repeats. Journal of the American Society for Horticultural Science, 126,205-209. |
| [14] | Chat J, Jáuregui B, Petit RJ, Nadot S (2004). Reticulate evolution in kiwifruit ( Actinidia, Actinidiaceae) identified by comparing their maternal and paternal phylogenies . American Journal of Botany, 915,736-747. |
| [15] | Cho YG, Ishii T, Temnykh S, Chen X, Lipovich L, McCouch SR, Park WD, Ayres N, Cartinhour S (2000). Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice ( Oryza sativa L.) . Theoretical and Applied Genetics, 100,713-722. |
| [16] | Choler P, Erschbamer B, Tribsch A, Gielly L, Taberiet P (2004). Genetic introgression as a potential to widen a species' niche: insights from alpine Carex curvula. Proceedings of the National Academy of Sciences of the United States of America 1,171-176. |
| [17] | Cipriani G, Marrazzo MT, Marconi R, Cimato A, Testolin R (2002). Microsatellite markers isolated in olive ( Olea europaea L.) are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars . Theoretical and Applied Genetics, 104,223-228. |
| [18] | Cordeiro GM, Pan YB, Henry RJ (2003). Sugarcane microsatellites for the assessment of genetic diversity in sugarcane germplasm. Plant Science, 165,181-189. |
| [19] |
Crowhurst RN, Gardner RC (1991). A genome-specific repeat sequence from kiwifruit ( Actinidia deliciosa var.deliciosa) Theoretical and Applied Genetics, 81,71-78.
URL PMID |
| [20] | Cruzan MB (2005). Patterns of introgression across an expanding hybrid zone: analysing historical patterns of gene flow using nonequilibrium approaches. New Phytologist, 167,267-278. |
| [21] | Cui ZX (崔致学) (1993). Actinidia in China (中国猕猴桃). Shandong Science and Technology Press, Ji'nan, 6-14. (in Chinese) |
| [22] | Excoffier L, Smouse PE, Quattro JM (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondria DNA restriction data. Genetics, 131,479-491. |
| [23] | Ferguson AR, Huang HW (2006). Genetic resources of kiwifruit: domestication and breeding. Horticultural Reviews, 33,1-121. |
| [24] |
Fukunaga K, Hill J, Vigouroux Y, Matsuoka Y, Sanchez J, Liu KJ, Buckler ES, Doebley J (2005). Genetic diversity and population structure of teosinte. Genetics, 169,2241-2254.
URL PMID |
| [25] | Gross R, Gum B, Reiter R, Kühn R (2004). Genetic introgression between Arctic charr ( Salvelinus alpinus) and brook trout ( Salvelinus fontinalis) in Bavarian hatchery stocks inferred from nuclear and mitochondrial DNA markers . Aquaculture International, 12,19-32. |
| [26] |
Gupta PK, Rustgi S, Sharma S, Singh R, Kumar N, Balyan HS (2003). Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Molecular Genetics and Genomics, 270,315-323.
URL PMID |
| [27] | Hamrick JL, Godt MJW (1989). Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS eds. Plant Population Genetics, Breeding, and Genetic Resources. Sinauer Associates Inc., Sunderland, 43-63. |
| [28] | Hamrick JL, Godt MJW (1996). Effects of life history traits on genetic diversity in plant species. Philosophical Transactions: Biological Sciences, 351,1291-1298. |
| [29] | Hokanson SC, Szewe-McFadden AK, Lamboy WF, McFerson JR (1998). Microsatellite (SSR) markers reveal genetic identities, genetic diversity and relationships in a Malus × domestica borkh Core subset collection . Theoretical and Applied Genetics, 97,671-683. |
| [30] | Huang HW (黄宏文), Gong JJ (龚俊杰), Wang SM (王圣梅), He ZC (何子灿), Zhang ZH (张忠慧), Li JQ (李建强) (2000). Genetic diversity in the genus Actinidia. Chinese Biodiversity (生物多样性), 8,1-12. (in Chinese with English abstract) |
| [31] | Huang HW, Dane F, Wang ZR, Jiang ZW, Huang RH, Wang SM (1997). Isozyme inheritance and variation in Actinidia. Heredity, 78,328-336. |
| [32] | Huang HW, Li JQ, Lang P, Wang SM (1999). Systematic relationships in Actinidia as revealed by cluster analysis of digitized morphological descriptors . Acta Horticulturae, 498,71-78. |
| [33] | Huang HW, Li ZZ, Li JQ, Kubisiak TL, Layne DR (2002). Phylogenetic relationships in Actinidia as revealed by RAPD analysis . Journal of the American Society for Horticultural Science, 127,759-766. |
| [34] | Huang WG, Cipriani G, Morgante M, Testolin R (1998). Microsatellite DNA in Actinidia chinensis: isolatin, characterization, and homology in related species . Theoretical and Applied Genetics, 97,1269-1278. |
| [35] | Ishihama F, Ueno S, Tsumura Y, Washitani I (2005). Gene flow and inbreeding depression inferred from fine-scale genetic structure in an endangered heterostylous perennial, Primula sieboldii. Molecular Ecology, 14,983-990. |
| [36] |
JakseJ, SatovicZ, JavornikB (2004). Microsatellite variability among wild and cultivated hops ( Humulus lupulus L. ) . Genome, 47,889-899.
DOI URL PMID |
| [37] |
Lehmann T, Hawley WA, Collins FH (1996). An evaluation of evolutionary constraints on microsatellite loci using null alleles. Genetics, 144,1155-1163.
URL PMID |
| [38] |
Lexer C, Kremer A, Petit RJ (2006). Shared alleles in sympatric oaks: recurrent gene flow is a more parsimonious explanation than ancestral polymorphism. Molecular Ecology, 15,2007-2012.
URL PMID |
| [39] | Li JQ, Huang HW, Sang T (2002). Molecular phylogeny and infrageneric classification of Actinidia (Actinidiaceae) . Systematic Botany, 27,408-415. |
| [40] | Li Q (栗琪), Li ZZ (李作洲), Huang HW (黄宏文) (2004). Preliminary study on SSR analysis in natural populations of Actinidia. Journal of Wuhan Botanical Research (武汉植物学研究), 22,175-178. (in Chinese with English abstract) |
| [41] | Li ZZ, Huang HW, Jiang ZW, Li JQ, Kubisiak TL (2003). Phylogenetic relationships in Actinidia as revealed by RAPDs and PCR-RFLPs of mtDNA . Acta Horticulturae, 610,387-396. |
| [42] | Liang CF (梁畴芬) (1975). Classification of Actinidia chinensis planch . Acta Phytotaxonomica Sinica (植物分类学报), 13,32-35. (in Chinese) |
| [43] | Liang CF (梁畴芬) (1983). On the distribution of Actinidias. Guihaia (广西植物), 3,229-248. (in Chinese with English abstract) |
| [44] | Liang CF (梁畴芬), Ferguson AR (1984). Emendation of the latin name of Actinidia chinensis pl. var. Hispida c.f.liang . Guihaia (广西植物), 4,181-182. (in Chinese with English abstract) |
| [45] | Llopart A, Lachaise D, Coyne JA (2005). Multilocus analysis of introgression between two sympatric sister species of Drosophila: Drosophila yakuba and D. santomea. Genetics, 171,197-210. |
| [46] | Mahy G, Bruederle LP, Connors B, van Hofwegen M, Vorsa N (2000). Allozyme evidence for genetic autopolyploidy and high genetic diversity in tetraploid cranberry, Vaccinium oxycoccos (Ericaceae). American Journal of Botany, 87,1882-1889. |
| [47] | Matus IA, Hayes PM (2002). Genetic diversity in three groups of barleys germplasm assessed by simple sequence repeats. Genome, 45,1095-1106. |
| [48] | Moody ME, Mueller LD, Soltis DE (1993). Genetic variation and random genetic drift in autotetraploid populations. Genetics, 134,649-657. |
| [49] |
Muir G, Schløtterer C (2005). Evidence for shared ancestral polymorphism rather than recurrent gene flow at microsatellite loci differentiating two hybridizing oaks ( Quercus spp. ) . Molecular Ecology, 14,549-561.
URL PMID |
| [50] | Nei M (1987). Molecular Evolutionary Genetics. Columbia University Press, New York. |
| [51] |
Nybom H (2004). Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology, 13,1143-1155.
URL PMID |
| [52] |
Ogden R, Thorpe RS (2002). Molecular evidence for ecological speciation in tropical habitats. Proceedings of the National Academy of Sciences of the United States of America, 99,13612-13615.
URL PMID |
| [53] | Randi E, Lucchini V (2002). Detecting rare introgression of domestic dog genes into wild wolf ( Canis lupus) populations by Bayesian admixture analyses of microsatellite variation . Conservation Genetics, 3,31-45. |
| [54] | Rieseberg LH, Carney SE (1998). Tansley review No. 102 plant hybridization. New Phytologist, 140,599-624. |
| [55] | Rogers SO, Bendich AJ (1985). Extraction of DNA from milligram amount of fresh, herbarium and mummified plant tissues. Plant Molecular Biology, 5,69-76. |
| [56] | Rohlf FJ (1994). NTSYS-pc, Numerical Taxonomy and Multivariate Analysis System Version 2.1 [CP/DK]. Exeter Ltd., Setauket, New York. |
| [57] |
Rongwen J, Akkaya MS, Bhagwat AA, Lavi U, Cregan PB (1995). The use of microsatelllite DNA markers for soybean genotype identification. Theoretical and Applied Genetics, 90,43-48.
URL PMID |
| [58] |
Rungis D, Bérubé Y, Zhang J, Ralph S, Ritland CE, Ellis BE, Douglas C, Bohlmann J, Ritland K (2004). Robust simple sequence repeat markers for spruce ( Picea spp.) from expressed sequence tags . Theoretical and Applied Genetics, 109,1283-1294.
DOI URL PMID |
| [59] | Sanguinetti CJ, Neto ED, Simpson AJG (1994). Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques, 17,915-919. |
| [60] | Schneider S, Roessli D, Excoffier L (2000). ARLEQUIN: A Software for Population Genetic Data Analysis, Version 2.000. Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva, Geneva. |
| [61] |
Slatkin M, Barton NH (1989). A comparison of three indirect methods for estimating average levels of gene flow. Evolution, 43,1349-1368.
DOI URL PMID |
| [62] |
Stehlik I, Holderegger R (2000). Spatial genetic structure and clonal diversity of Anemone nemorosa in late successional deciduous woodlands of Central Europe . Journal of Ecology, 88,424-435.
URL PMID |
| [63] |
Streiff R, Veyrier R, Audiot P, Meusnier S, Brouat C (2005). Introgression in natural populations of bioindicators: a case study of Carabus splendens and Carabus punctatoauratus. Molecular Ecology, 14,3775-3786.
DOI URL PMID |
| [64] | Testolin R, Ferguson AR (1997). Isozyme polymorphism in the genus Actinidia and the origin of the kiwifruit genome . Systematic Botany, 22,685-700. |
| [65] |
Tovar-Sánchez E, Oyama K (2004). Natural hybridization and hybrid zones between Quercus crassifolia and Quercus crassipes (Fagaceae) in Mexico: morphological and molecular evidence . American Journal of Botany, 91,1352-1363.
URL PMID |
| [66] | van Droogenbroeck B, Kyndt T, Romeijn-Peeters E, van Thuyne W, Goetghebeur P, Romero-Motochi JP, Gheysen G (2006). Evidence of natural hybridization and introgression between Vasconcellea species (Caricaceae) from southern Ecuador revealed by chloroplast, mitochondrial and nuclear DNA markers . Annals of Botany, 97,193-805. |
| [67] | Wang SM (王圣梅), Huang RH (黄仁煌), Wu XW (吴显维), Kang N (康宁) (1994). Studies on Actinidia breeding by species hybridization . Journal of Fruit Science (果树科学), 11,23-26. (in Chinese with English abstract) |
| [68] | Weising K, Fung RWM, Keeling DJ, Atkinson RG, Gardner RC (1996). Characterization of microsatellites from Actinidia chinensis. Molecular Breeding, 2,117-131. |
| [69] | Wright S (1943). Isoltion by distance. Genetics, 28,114-138. |
| [70] | Zhang ZY (张芝玉) (1983). A report on the chromosome numbers of 2 varieties of Actinidia chinensis Planch . Acta Phytotaxonomica Sinica (植物分类学报), 21,161-163. (in Chinese with English abstract) |
| [71] | Zhen YQ, Li ZZ, Huang HW, Wang Y (2004). Molecular characterization of kiwifruit ( Actinidia) cultivars and selections using SSR markers . Journal of the American Society for Horticultural Science, 129,374-382. |
| [72] | Zou YP (邹喻苹), Ge S (葛颂), Wang XD (王晓东) (2001). Molecular Markers of Systematic and Evolutionary Botany (系统与进化植物学中的分子标记). Science Press, Beijing,68-107. (in Chinese) |
| [1] | 陈天翌, 娄安如. 青藏高原东侧白桦种群的遗传多样性与遗传结构[J]. 植物生态学报, 2022, 46(5): 561-568. |
| [2] | 杜军, 王文, 何志斌, 陈龙飞, 蔺鹏飞, 朱喜, 田全彦. 祁连山青海云杉物候表型的空间分异及其内在机制[J]. 植物生态学报, 2021, 45(8): 834-843. |
| [3] | 张新新, 王茜, 胡颖, 周玮, 陈晓阳, 胡新生. 植物边缘种群遗传多样性研究进展[J]. 植物生态学报, 2019, 43(5): 383-395. |
| [4] | 张俪文, 韩广轩. 植物遗传多样性与生态系统功能关系的研究进展[J]. 植物生态学报, 2018, 42(10): 977-989. |
| [5] | 杨雪, 申俊芳, 赵念席, 高玉葆. 不同基因型羊草数量性状的可塑性及遗传分化[J]. 植物生态学报, 2017, 41(3): 359-368. |
| [6] | 王锦楠, 陈进福, 陈武生, 周新洋, 许东, 李际红, 亓晓. 柴达木地区野生黑果枸杞种群遗传多样性的AFLP分析[J]. 植物生态学报, 2015, 39(10): 1003-1011. |
| [7] | 刘军, 姜景民, 邹军, 徐金良, 沈汉, 刁松峰. 中国特有濒危树种毛红椿核心和边缘居群的遗传多样性[J]. 植物生态学报, 2013, 37(1): 52-60. |
| [8] | 张炜, 罗建勋, 辜云杰, 胡庭兴. 西南地区麻疯树天然种群遗传多样性的等位酶变异[J]. 植物生态学报, 2011, 35(3): 330-336. |
| [9] | 魏源, 王世杰, 刘秀明, 黄天志. 不同喀斯特小生境中土壤丛枝菌根真菌的遗传多样性[J]. 植物生态学报, 2011, 35(10): 1083-1090. |
| [10] | 张云红, 侯艳, 娄安如. 华北地区小丛红景天种群的AFLP遗传多样性[J]. 植物生态学报, 2010, 34(9): 1084-1094. |
| [11] | 刘伟, 王曦, 干友民, 黄林凯, 谢文刚, 苗佳敏. 高山嵩草种群在放牧干扰下遗传多样性的变化[J]. 植物生态学报, 2009, 33(5): 966-973. |
| [12] | 张冬梅, 孙佩光, 沈熙环, 茹广欣. 油松种子园自由授粉与控制授粉种子父本分析[J]. 植物生态学报, 2009, 33(2): 302-310. |
| [13] | 陈良华, 胡庭兴, 张帆, 李国和. 用AFLP技术分析四川核桃资源的遗传多样性[J]. 植物生态学报, 2008, 32(6): 1362-1372. |
| [14] | 周会平, 陈进, 张寿洲. 具混合繁殖策略的草本植物异果舞花姜的居群遗传结构[J]. 植物生态学报, 2008, 32(4): 751-759. |
| [15] | 严茂粉, 李向华, 王克晶. 北京地区野生大豆种群SSR标记的遗传多样性评价[J]. 植物生态学报, 2008, 32(4): 938-950. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
Copyright © 2022 版权所有 《植物生态学报》编辑部
地址: 北京香山南辛村20号, 邮编: 100093
Tel.: 010-62836134, 62836138; Fax: 010-82599431; E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
备案号: 京ICP备16067583号-19