

植物生态学报 ›› 2013, Vol. 37 ›› Issue (1): 52-60.DOI: 10.3724/SP.J.1258.2013.00006
所属专题: 生物多样性
刘军1, 姜景民1,*( ), 邹军2, 徐金良3, 沈汉3, 刁松峰1
), 邹军2, 徐金良3, 沈汉3, 刁松峰1
收稿日期:2012-08-27
接受日期:2012-11-28
出版日期:2013-08-27
发布日期:2013-01-15
通讯作者:
姜景民
作者简介:*(E-mail:jiang_jingmin@163.com)基金资助:
LIU Jun1, JIANG Jing-Min1,*( ), ZOU Jun2, XU Jin-Liang3, SHEN Han3, DIAO Song-Feng1
), ZOU Jun2, XU Jin-Liang3, SHEN Han3, DIAO Song-Feng1
Received:2012-08-27
Accepted:2012-11-28
Online:2013-08-27
Published:2013-01-15
Contact:
JIANG Jing-Min
摘要:
选用8对多态的微卫星引物, 研究了毛红椿(Toona ciliata var. pubescens)核心居群和边缘居群的遗传多样性。研究结果显示, 边缘居群的观察等位基因数和有效等位基因数并不比核心居群低, 甚至更高。普通广布和稀有地方等位基因在所有居群中均有分布, 而普通地方和稀有广布等位基因分别仅在5个和3个居群中有分布。从4种类型等位基因总数上看, 边缘居群高于核心居群, 特别是边缘居群的稀有地方等位基因数量明显多于核心居群。边缘居群的平均期望杂合度和观察杂合度都高于核心居群。核心居群间的基因分化系数为0.1520, 边缘居群间为0.3045, 边缘居群与核心居群的遗传分化差异达到显著水平。核心居群间基因流大于1, 而边缘居群间基因流小于1, 说明核心居群间基因交流频繁, 边缘居群由于片段化和地形影响, 基因流较小。Mantel检验结果显示, 居群间遗传距离与地理距离的相关性不显著, 说明地理距离对毛红椿居群遗传分化的影响不显著。
刘军, 姜景民, 邹军, 徐金良, 沈汉, 刁松峰. 中国特有濒危树种毛红椿核心和边缘居群的遗传多样性. 植物生态学报, 2013, 37(1): 52-60. DOI: 10.3724/SP.J.1258.2013.00006
LIU Jun, JIANG Jing-Min, ZOU Jun, XU Jin-Liang, SHEN Han, DIAO Song-Feng. Genetic diversity of central and peripheral populations of Toona ciliata var. pubescens, an endangered tree species endemic to China. Chinese Journal of Plant Ecology, 2013, 37(1): 52-60. DOI: 10.3724/SP.J.1258.2013.00006
| 居群 Population | 所属省份 Province | 类型 Type | 经度 Longitude | 纬度 Latitude | 海拔高度 Altitude (m) | 采样数量 Sampled number |
|---|---|---|---|---|---|---|
| 宜丰 | 江西 | 核心居群 | 114°29′-114°45′ E | 28°30′-28°40′ N | 220-475 | 65 |
| Yifeng (YF) | Jiangxi | Central population | ||||
| 泾县 | 安徽 | 核心居群 | 118°35′-118°36′ E | 30°31′-30°34′ N | 270-497 | 48 |
| Jingxian (JX) | Anhui | Central population | ||||
| 仙居 | 浙江 | 核心居群 | 120°32′-120°56′ E | 28°48′-28°56′ N | 600-820 | 27 |
| Xianju (XJ) | Zhejiang | Central population | ||||
| 遂昌 | 浙江 | 核心居群 | 119°12′-119°23′ E | 28°30′-28°36′ N | 510-1 220 | 25 |
| Suichang (SC) | Zhejiang | Central population | ||||
| 宾川 | 云南 | 边缘居群 | 100°16′-101°23′ E | 25°02′-25°22′ N | 1 404-1 820 | 60 |
| Binchuan (BC) | Yunnan | Peripheral population | ||||
| 元谋 | 云南 | 边缘居群 | 101°49′-101°52′ E | 25°17′-25°40′ N | 1 112-1 230 | 30 |
| Yuanmou (YM) | Yunnan | Peripheral population | ||||
| 武定 | 云南 | 边缘居群 | 102°08′-102°12′ E | 25°47′-25°51′ N | 1 405-1 802 | 29 |
| Wuding (WD) | Yunnan | Peripheral population | ||||
| 师宗 | 云南 | 边缘居群 | 103°42′-104°34′ E | 24°21′-25°00′ N | 812-912 | 84 |
| Shizong (SZ) | Yunnan | Peripheral population | ||||
| 册亨 | 贵州 | 边缘居群 | 105°40′-105°52′ E | 24°36′-24°55′ N | 620-810 | 24 |
| Ceheng (CH) | Guizhou | Peripheral population |
表1 毛红椿9个天然居群地理位置和采样数量
Table 1 Geographic place and individuals sampled for nine natural populations of Toona ciliata var. pubescens
| 居群 Population | 所属省份 Province | 类型 Type | 经度 Longitude | 纬度 Latitude | 海拔高度 Altitude (m) | 采样数量 Sampled number |
|---|---|---|---|---|---|---|
| 宜丰 | 江西 | 核心居群 | 114°29′-114°45′ E | 28°30′-28°40′ N | 220-475 | 65 |
| Yifeng (YF) | Jiangxi | Central population | ||||
| 泾县 | 安徽 | 核心居群 | 118°35′-118°36′ E | 30°31′-30°34′ N | 270-497 | 48 |
| Jingxian (JX) | Anhui | Central population | ||||
| 仙居 | 浙江 | 核心居群 | 120°32′-120°56′ E | 28°48′-28°56′ N | 600-820 | 27 |
| Xianju (XJ) | Zhejiang | Central population | ||||
| 遂昌 | 浙江 | 核心居群 | 119°12′-119°23′ E | 28°30′-28°36′ N | 510-1 220 | 25 |
| Suichang (SC) | Zhejiang | Central population | ||||
| 宾川 | 云南 | 边缘居群 | 100°16′-101°23′ E | 25°02′-25°22′ N | 1 404-1 820 | 60 |
| Binchuan (BC) | Yunnan | Peripheral population | ||||
| 元谋 | 云南 | 边缘居群 | 101°49′-101°52′ E | 25°17′-25°40′ N | 1 112-1 230 | 30 |
| Yuanmou (YM) | Yunnan | Peripheral population | ||||
| 武定 | 云南 | 边缘居群 | 102°08′-102°12′ E | 25°47′-25°51′ N | 1 405-1 802 | 29 |
| Wuding (WD) | Yunnan | Peripheral population | ||||
| 师宗 | 云南 | 边缘居群 | 103°42′-104°34′ E | 24°21′-25°00′ N | 812-912 | 84 |
| Shizong (SZ) | Yunnan | Peripheral population | ||||
| 册亨 | 贵州 | 边缘居群 | 105°40′-105°52′ E | 24°36′-24°55′ N | 620-810 | 24 |
| Ceheng (CH) | Guizhou | Peripheral population |
| 位点 Loci | GenBank Accession No. | 引物序列 Primer sequence (5′-3′) | 重复序列 Repeat motif | 退火温度 Annealing temperature (℃) | 等位基因大小 Allele size (bp) |
|---|---|---|---|---|---|
| Tc01 | DQ778303 | F: TCAATGCAATTTAGGAGGAA | (GA)8 | 52 | 240-291 |
| R: TGCTTGTTGAACCCTGTG | |||||
| Tc02 | DQ453904 | F: TAGGAAAGGCAAGGTGGG | (AG)14 | 55 | 109-120 |
| R: GGGTGGTCGATGAGGGTT | |||||
| Tc03 | DQ453907 | F: GATTACGCCAGGCAAACG | (CT)6 | 55 | 230-320 |
| R: TTGAATATGGGAGAAGGT | |||||
| Tc04 | DQ453906 | F: GAAACCAGCAGGCAGAGC | (AG)10 | 55 | 110-230 |
| R: GAAGAAGGGTGAGCGAGA | |||||
| Tc05 | DQ453905 | F: AGTAATAGCCTGTAGAGCAG | (AG)13 | 55 | 120-242 |
| R: AGAGTGGGGTGGTCGATGAG | |||||
| Tc06 | DQ453903 | F: GACTCGTGACACTTAGCCTGTA | (TTTCTC)7 | 55 | 121-231 |
| R: CTGGCGTAATCATGGTCATAC | |||||
| Tc07 | DQ453912 | F: ATGGATGAGTGTGCGATAGG | (TC)7 | 55 | 182-280 |
| R: TGTGATGTAGGAGTCTGAAC | |||||
| Tc08 | DQ453914 | F: TGTCTCAGTTTATGCTGGCGT | (TC)8 | 55 | 170-260 |
| R: CTGCCCAATCAACAAGAG |
表2 毛红椿基因组8个微卫星位点的PCR扩增引物
Table 2 PCR primers for amplifying eight SSR loci in Toona ciliata var. pubescens
| 位点 Loci | GenBank Accession No. | 引物序列 Primer sequence (5′-3′) | 重复序列 Repeat motif | 退火温度 Annealing temperature (℃) | 等位基因大小 Allele size (bp) |
|---|---|---|---|---|---|
| Tc01 | DQ778303 | F: TCAATGCAATTTAGGAGGAA | (GA)8 | 52 | 240-291 |
| R: TGCTTGTTGAACCCTGTG | |||||
| Tc02 | DQ453904 | F: TAGGAAAGGCAAGGTGGG | (AG)14 | 55 | 109-120 |
| R: GGGTGGTCGATGAGGGTT | |||||
| Tc03 | DQ453907 | F: GATTACGCCAGGCAAACG | (CT)6 | 55 | 230-320 |
| R: TTGAATATGGGAGAAGGT | |||||
| Tc04 | DQ453906 | F: GAAACCAGCAGGCAGAGC | (AG)10 | 55 | 110-230 |
| R: GAAGAAGGGTGAGCGAGA | |||||
| Tc05 | DQ453905 | F: AGTAATAGCCTGTAGAGCAG | (AG)13 | 55 | 120-242 |
| R: AGAGTGGGGTGGTCGATGAG | |||||
| Tc06 | DQ453903 | F: GACTCGTGACACTTAGCCTGTA | (TTTCTC)7 | 55 | 121-231 |
| R: CTGGCGTAATCATGGTCATAC | |||||
| Tc07 | DQ453912 | F: ATGGATGAGTGTGCGATAGG | (TC)7 | 55 | 182-280 |
| R: TGTGATGTAGGAGTCTGAAC | |||||
| Tc08 | DQ453914 | F: TGTCTCAGTTTATGCTGGCGT | (TC)8 | 55 | 170-260 |
| R: CTGCCCAATCAACAAGAG |
| 位点 Loci | 所有居群 All populations | 核心居群 Central population | 边缘居群 Peripheral population | |||||
|---|---|---|---|---|---|---|---|---|
| 观察等位基因数 Na | 有效等位基因数 Ne | 观察等位基因数 Na | 有效等位基因数 Ne | 观察等位基因数 Na | 有效等位基因数 Ne | |||
| Tc01 | 6.00 | 3.37 | 3.00 | 1.98 | 5.00 | 3.64 | ||
| Tc02 | 6.00 | 1.60 | 3.00 | 1.73 | 6.00 | 1.48 | ||
| Tc03 | 6.00 | 2.57 | 4.00 | 2.94 | 6.00 | 2.27 | ||
| Tc04 | 8.00 | 3.42 | 4.00 | 3.00 | 6.00 | 2.57 | ||
| Tc05 | 8.00 | 3.36 | 3.00 | 2.93 | 8.00 | 2.57 | ||
| Tc06 | 8.00 | 3.87 | 6.00 | 3.23 | 8.00 | 4.25 | ||
| Tc07 | 8.00 | 3.44 | 7.00 | 2.58 | 8.00 | 4.06 | ||
| Tc08 | 6.00 | 2.45 | 6.00 | 1.82 | 4.00 | 2.09 | ||
| 平均值 Mean | 7.00 | 3.01 | 4.75 | 2.53 | 6.38 | 2.87 | ||
| 标准偏差 SD | 1.07 | 0.74 | 1.58 | 0.59 | 1.51 | 1.00 | ||
表3 毛红椿天然居群观察等位基因和有效等位基因数量
Table 3 Observed (Na) and effective number of alleles (Ne) in populations of Toona ciliata var. pubescens
| 位点 Loci | 所有居群 All populations | 核心居群 Central population | 边缘居群 Peripheral population | |||||
|---|---|---|---|---|---|---|---|---|
| 观察等位基因数 Na | 有效等位基因数 Ne | 观察等位基因数 Na | 有效等位基因数 Ne | 观察等位基因数 Na | 有效等位基因数 Ne | |||
| Tc01 | 6.00 | 3.37 | 3.00 | 1.98 | 5.00 | 3.64 | ||
| Tc02 | 6.00 | 1.60 | 3.00 | 1.73 | 6.00 | 1.48 | ||
| Tc03 | 6.00 | 2.57 | 4.00 | 2.94 | 6.00 | 2.27 | ||
| Tc04 | 8.00 | 3.42 | 4.00 | 3.00 | 6.00 | 2.57 | ||
| Tc05 | 8.00 | 3.36 | 3.00 | 2.93 | 8.00 | 2.57 | ||
| Tc06 | 8.00 | 3.87 | 6.00 | 3.23 | 8.00 | 4.25 | ||
| Tc07 | 8.00 | 3.44 | 7.00 | 2.58 | 8.00 | 4.06 | ||
| Tc08 | 6.00 | 2.45 | 6.00 | 1.82 | 4.00 | 2.09 | ||
| 平均值 Mean | 7.00 | 3.01 | 4.75 | 2.53 | 6.38 | 2.87 | ||
| 标准偏差 SD | 1.07 | 0.74 | 1.58 | 0.59 | 1.51 | 1.00 | ||
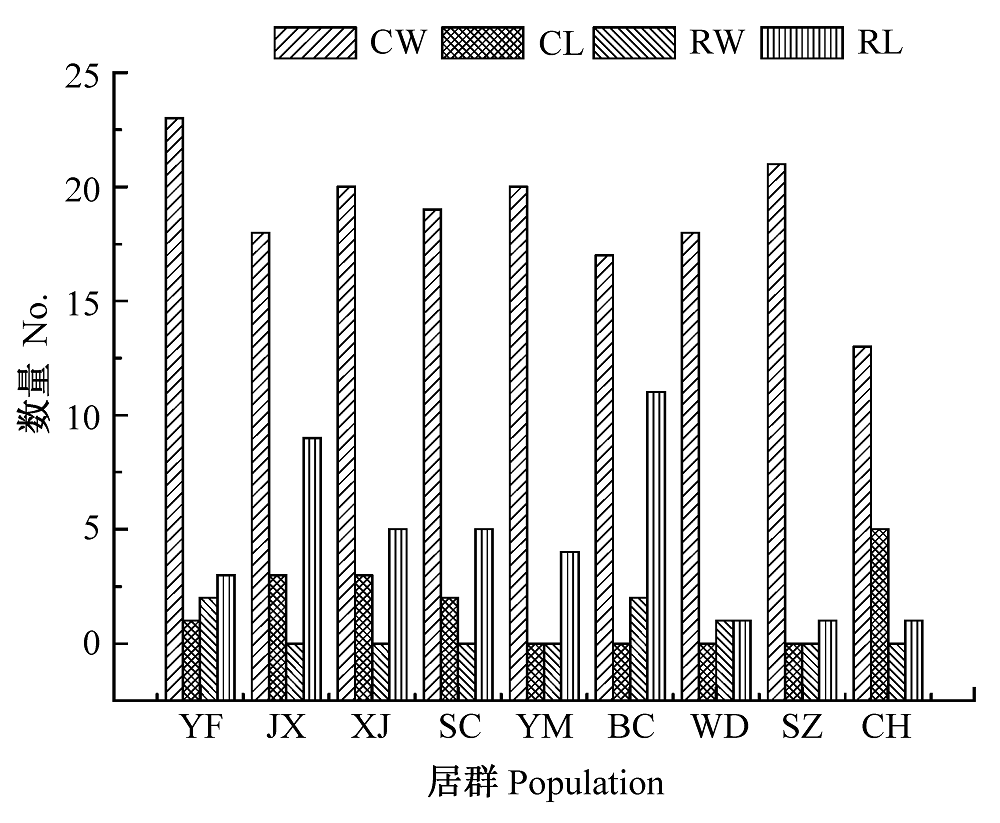
图1 毛红椿9个天然居群不同类型等位基因分布图。CL, 普通地方等位基因; CW, 普通广布等位基因; RL, 稀有地方等位基因; RW, 稀有广布等位基因。居群同表1。
Fig. 1 Distribution of different types of alleles in nine natural populations of Toona ciliata var. pubescens. CL, common local allele; CW, common wide allele; RL, rare local allele; RW, rare wide allele. Population see Table 1.
| 位点 Loci | 观察杂合度 HO | 期望杂合度 HE | Nei’s期望杂合度 Nei’s HE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 所有居群 All populations | 核心居群 Central population | 边缘居群Peripheral population | 所有居群 All populations | 核心居群 Central population | 边缘居群Peripheral population | 所有居群 All populations | 核心居群 Central population | 边缘居群 Peripheral population | |||
| Tc01 | 0.591 1 | 0.222 9 | 0.845 8 | 0.704 4 | 0.496 4 | 0.726 9 | 0.703 5 | 0.494 8 | 0.725 3 | ||
| Tc02 | 0.354 2 | 0.560 5 | 0.211 5 | 0.376 2 | 0.425 8 | 0.324 3 | 0.375 7 | 0.424 4 | 0.323 6 | ||
| Tc03 | 0.492 2 | 0.598 7 | 0.418 5 | 0.611 3 | 0.661 5 | 0.561 1 | 0.610 5 | 0.659 4 | 0.559 8 | ||
| Tc04 | 0.783 9 | 0.808 9 | 0.766 5 | 0.709 0 | 0.669 5 | 0.612 1 | 0.708 1 | 0.667 4 | 0.610 7 | ||
| Tc05 | 0.504 2 | 0.562 9 | 0.461 5 | 0.703 1 | 0.661 4 | 0.612 8 | 0.702 2 | 0.659 2 | 0.611 3 | ||
| Tc06 | 0.765 9 | 0.708 0 | 0.814 8 | 0.742 6 | 0.693 0 | 0.767 3 | 0.741 3 | 0.690 5 | 0.765 0 | ||
| Tc07 | 0.522 4 | 0.344 8 | 0.657 9 | 0.710 1 | 0.614 2 | 0.755 9 | 0.709 0 | 0.612 0 | 0.753 9 | ||
| Tc08 | 0.673 0 | 0.472 4 | 0.808 5 | 0.592 6 | 0.452 2 | 0.522 2 | 0.591 7 | 0.450 4 | 0.520 8 | ||
| 平均 Mean | 0.585 9 | 0.534 9 | 0.623 1 | 0.643 7 | 0.584 2 | 0.610 3 | 0.642 7 | 0.582 3 | 0.608 8 | ||
表4 毛红椿核心和边缘居群8个微卫星位点观察杂合度和期望杂合度
Table 4 Observed heterozygosity (HO) and expected heterozygosity (HE) of eight microsatellite loci in central and peripheral population of Toona ciliata var. pubescens
| 位点 Loci | 观察杂合度 HO | 期望杂合度 HE | Nei’s期望杂合度 Nei’s HE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 所有居群 All populations | 核心居群 Central population | 边缘居群Peripheral population | 所有居群 All populations | 核心居群 Central population | 边缘居群Peripheral population | 所有居群 All populations | 核心居群 Central population | 边缘居群 Peripheral population | |||
| Tc01 | 0.591 1 | 0.222 9 | 0.845 8 | 0.704 4 | 0.496 4 | 0.726 9 | 0.703 5 | 0.494 8 | 0.725 3 | ||
| Tc02 | 0.354 2 | 0.560 5 | 0.211 5 | 0.376 2 | 0.425 8 | 0.324 3 | 0.375 7 | 0.424 4 | 0.323 6 | ||
| Tc03 | 0.492 2 | 0.598 7 | 0.418 5 | 0.611 3 | 0.661 5 | 0.561 1 | 0.610 5 | 0.659 4 | 0.559 8 | ||
| Tc04 | 0.783 9 | 0.808 9 | 0.766 5 | 0.709 0 | 0.669 5 | 0.612 1 | 0.708 1 | 0.667 4 | 0.610 7 | ||
| Tc05 | 0.504 2 | 0.562 9 | 0.461 5 | 0.703 1 | 0.661 4 | 0.612 8 | 0.702 2 | 0.659 2 | 0.611 3 | ||
| Tc06 | 0.765 9 | 0.708 0 | 0.814 8 | 0.742 6 | 0.693 0 | 0.767 3 | 0.741 3 | 0.690 5 | 0.765 0 | ||
| Tc07 | 0.522 4 | 0.344 8 | 0.657 9 | 0.710 1 | 0.614 2 | 0.755 9 | 0.709 0 | 0.612 0 | 0.753 9 | ||
| Tc08 | 0.673 0 | 0.472 4 | 0.808 5 | 0.592 6 | 0.452 2 | 0.522 2 | 0.591 7 | 0.450 4 | 0.520 8 | ||
| 平均 Mean | 0.585 9 | 0.534 9 | 0.623 1 | 0.643 7 | 0.584 2 | 0.610 3 | 0.642 7 | 0.582 3 | 0.608 8 | ||
| 居群 Population | 观察杂合度 HO | 期望杂合度 HE | Nei’s期望杂合度 Nei’s HE |
|---|---|---|---|
| 宜丰 YF | 0.581 1 | 0.492 0 | 0.488 0 |
| 泾县 JX | 0.486 2 | 0.502 3 | 0.496 4 |
| 仙居 XJ | 0.610 3 | 0.467 4 | 0.454 8 |
| 遂昌 SC | 0.501 9 | 0.514 0 | 0.502 9 |
| 元谋 YM | 0.669 2 | 0.603 0 | 0.589 9 |
| 宾川 BC | 0.759 6 | 0.502 5 | 0.497 7 |
| 武定 WD | 0.684 8 | 0.490 0 | 0.480 5 |
| 师宗 SZ | 0.592 6 | 0.430 4 | 0.427 6 |
| 册亨 CH | 0.214 3 | 0.435 7 | 0.426 6 |
| 平均值 Mean | 0.585 9 | 0.643 7 | 0.642 7 |
表5 毛红椿9个天然居群遗传多样性
Table 5 Genetic diversity in nine natural populations of Toona ciliata var. pubescens
| 居群 Population | 观察杂合度 HO | 期望杂合度 HE | Nei’s期望杂合度 Nei’s HE |
|---|---|---|---|
| 宜丰 YF | 0.581 1 | 0.492 0 | 0.488 0 |
| 泾县 JX | 0.486 2 | 0.502 3 | 0.496 4 |
| 仙居 XJ | 0.610 3 | 0.467 4 | 0.454 8 |
| 遂昌 SC | 0.501 9 | 0.514 0 | 0.502 9 |
| 元谋 YM | 0.669 2 | 0.603 0 | 0.589 9 |
| 宾川 BC | 0.759 6 | 0.502 5 | 0.497 7 |
| 武定 WD | 0.684 8 | 0.490 0 | 0.480 5 |
| 师宗 SZ | 0.592 6 | 0.430 4 | 0.427 6 |
| 册亨 CH | 0.214 3 | 0.435 7 | 0.426 6 |
| 平均值 Mean | 0.585 9 | 0.643 7 | 0.642 7 |
| 位点 Loci | Fis | Fit | FST | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 所有居群 All populations | 核心居群 Central population | 边缘居群 Peripheral population | 所有居群 All populations | 核心居群 Central population | 边缘居群 Peripheral population | 所有居群 All populations | 核心居群 Central population | 边缘居群 Peripheral population | |||
| Tc01 | -0.158 5 | 0.479 7 | -0.531 6 | 0.312 5 | 0.603 6 | 0.043 4 | 0.406 6 | 0.238 1 | 0.375 4 | ||
| Tc02 | -0.316 0 | -0.460 6 | -0.157 9 | 0.063 8 | -0.349 5 | 0.317 7 | 0.288 6 | 0.076 1 | 0.410 8 | ||
| Tc03 | 0.114 9 | -0.079 9 | 0.285 7 | 0.281 5 | 0.076 5 | 0.438 9 | 0.188 3 | 0.144 8 | 0.214 5 | ||
| Tc04 | -0.397 5 | -0.443 0 | -0.356 9 | -0.047 0 | -0.267 8 | -0.034 6 | 0.250 8 | 0.121 4 | 0.237 5 | ||
| Tc05 | -0.585 9 | -0.750 3 | -0.437 2 | 0.302 4 | 0.034 3 | 0.368 6 | 0.560 1 | 0.443 8 | 0.560 7 | ||
| Tc06 | -0.235 1 | -0.086 9 | -0.372 2 | 0.005 4 | -0.034 0 | 0.005 9 | 0.194 7 | 0.048 7 | 0.275 5 | ||
| Tc07 | 0.267 0 | 0.429 9 | 0.159 8 | 0.316 7 | 0.452 5 | 0.208 2 | 0.067 8 | 0.039 6 | 0.057 7 | ||
| Tc08 | -0.393 6 | -0.110 6 | -0.620 3 | 0.129 4 | -0.041 4 | 0.051 3 | 0.375 3 | 0.062 3 | 0.414 5 | ||
| 平均值 Mean | -0.163 1 | -0.108 5 | -0.209 9 | 0.175 0 | 0.060 0 | 0.158 6 | 0.290 7 | 0.152 0 | 0.304 5 | ||
表6 毛红椿核心和边缘居群F统计量
Table 6 Estimated F-statistics in central and peripheral populations of Toona ciliata var. pubescens
| 位点 Loci | Fis | Fit | FST | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 所有居群 All populations | 核心居群 Central population | 边缘居群 Peripheral population | 所有居群 All populations | 核心居群 Central population | 边缘居群 Peripheral population | 所有居群 All populations | 核心居群 Central population | 边缘居群 Peripheral population | |||
| Tc01 | -0.158 5 | 0.479 7 | -0.531 6 | 0.312 5 | 0.603 6 | 0.043 4 | 0.406 6 | 0.238 1 | 0.375 4 | ||
| Tc02 | -0.316 0 | -0.460 6 | -0.157 9 | 0.063 8 | -0.349 5 | 0.317 7 | 0.288 6 | 0.076 1 | 0.410 8 | ||
| Tc03 | 0.114 9 | -0.079 9 | 0.285 7 | 0.281 5 | 0.076 5 | 0.438 9 | 0.188 3 | 0.144 8 | 0.214 5 | ||
| Tc04 | -0.397 5 | -0.443 0 | -0.356 9 | -0.047 0 | -0.267 8 | -0.034 6 | 0.250 8 | 0.121 4 | 0.237 5 | ||
| Tc05 | -0.585 9 | -0.750 3 | -0.437 2 | 0.302 4 | 0.034 3 | 0.368 6 | 0.560 1 | 0.443 8 | 0.560 7 | ||
| Tc06 | -0.235 1 | -0.086 9 | -0.372 2 | 0.005 4 | -0.034 0 | 0.005 9 | 0.194 7 | 0.048 7 | 0.275 5 | ||
| Tc07 | 0.267 0 | 0.429 9 | 0.159 8 | 0.316 7 | 0.452 5 | 0.208 2 | 0.067 8 | 0.039 6 | 0.057 7 | ||
| Tc08 | -0.393 6 | -0.110 6 | -0.620 3 | 0.129 4 | -0.041 4 | 0.051 3 | 0.375 3 | 0.062 3 | 0.414 5 | ||
| 平均值 Mean | -0.163 1 | -0.108 5 | -0.209 9 | 0.175 0 | 0.060 0 | 0.158 6 | 0.290 7 | 0.152 0 | 0.304 5 | ||
| 居群 Population | 宜丰 YF | 泾县 JX | 仙居 XJ | 遂昌 SC | 元谋 YM | 宾川 BC | 武定 WD | 师宗 SZ | 册亨 CH |
|---|---|---|---|---|---|---|---|---|---|
| 宜丰 YF | 0.000 0 | 0.617 8 | 1.442 6 | 0.547 7 | 0.754 4 | 0.650 9 | 46.046 3 | 12.440 4 | 0.261 1 |
| 泾县 JX | 0.288 1** | 0.000 0 | 0.869 1 | 0.649 9 | 0.650 9 | 0.567 3 | 0.638 1 | 0.691 3 | 0.308 5 |
| 仙居 XJ | 0.147 7 ns | 0.223 4** | 0.000 0 | 0.988 9 | 1.471 8 | 1.013 9 | 1.311 5 | 2.124 2 | 0.301 3 |
| 遂昌 SC | 0.313 4 ns | 0.277 8** | 0.201 8** | 0.000 0 | 11.769 2 | 3.902 8 | 0.474 0 | 0.662 7 | 0.190 5 |
| 元谋 YM | 0.248 9** | 0.277 5** | 0.145 2** | 0.020 8** | 0.000 0 | 7.866 9 | 0.666 4 | 0.964 8 | 0.220 3 |
| 宾川 BC | 0.277 5** | 0.305 9** | 0.197 8** | 0.060 2** | 0.030 8** | 0.000 0 | 0.587 2 | 0.702 7 | 0.179 8 |
| 武定 WD | 0.005 4** | 0.281 5** | 0.160 1** | 0.345 3** | 0.272 8** | 0.298 6** | 0.000 0 | 4.770 1 | 0.257 4 |
| 师宗 SZ | 0.019 7** | 0.265 6** | 0.105 3** | 0.273 9** | 0.205 8** | 0.262 4** | 0.049 8** | 0.000 0 | 0.255 3 |
| 册亨 CH | 0.489 1ns | 0.447 6ns | 0.453 5ns | 0.567 6ns | 0.531 6ns | 0.581 6ns | 0.492 7ns | 0.494 8ns | 0.000 0 |
表7 毛红椿居群间的基因流(上三角)和基因分化系数(下三角)
Table 7 Gene flow value (upper triangle) and gene differentiation coefficient value (lower triangle) among populations of Toona ciliata var. pubescens
| 居群 Population | 宜丰 YF | 泾县 JX | 仙居 XJ | 遂昌 SC | 元谋 YM | 宾川 BC | 武定 WD | 师宗 SZ | 册亨 CH |
|---|---|---|---|---|---|---|---|---|---|
| 宜丰 YF | 0.000 0 | 0.617 8 | 1.442 6 | 0.547 7 | 0.754 4 | 0.650 9 | 46.046 3 | 12.440 4 | 0.261 1 |
| 泾县 JX | 0.288 1** | 0.000 0 | 0.869 1 | 0.649 9 | 0.650 9 | 0.567 3 | 0.638 1 | 0.691 3 | 0.308 5 |
| 仙居 XJ | 0.147 7 ns | 0.223 4** | 0.000 0 | 0.988 9 | 1.471 8 | 1.013 9 | 1.311 5 | 2.124 2 | 0.301 3 |
| 遂昌 SC | 0.313 4 ns | 0.277 8** | 0.201 8** | 0.000 0 | 11.769 2 | 3.902 8 | 0.474 0 | 0.662 7 | 0.190 5 |
| 元谋 YM | 0.248 9** | 0.277 5** | 0.145 2** | 0.020 8** | 0.000 0 | 7.866 9 | 0.666 4 | 0.964 8 | 0.220 3 |
| 宾川 BC | 0.277 5** | 0.305 9** | 0.197 8** | 0.060 2** | 0.030 8** | 0.000 0 | 0.587 2 | 0.702 7 | 0.179 8 |
| 武定 WD | 0.005 4** | 0.281 5** | 0.160 1** | 0.345 3** | 0.272 8** | 0.298 6** | 0.000 0 | 4.770 1 | 0.257 4 |
| 师宗 SZ | 0.019 7** | 0.265 6** | 0.105 3** | 0.273 9** | 0.205 8** | 0.262 4** | 0.049 8** | 0.000 0 | 0.255 3 |
| 册亨 CH | 0.489 1ns | 0.447 6ns | 0.453 5ns | 0.567 6ns | 0.531 6ns | 0.581 6ns | 0.492 7ns | 0.494 8ns | 0.000 0 |
| 居群 Population | 宜丰 YF | 泾县 JX | 仙居 XJ | 遂昌 SC | 元谋 YM | 宾川 BC | 武定 WD | 师宗 SZ | 册亨 CH |
|---|---|---|---|---|---|---|---|---|---|
| 宜丰 YF | 0.000 0 | 466.550 4 | 669.412 6 | 522.624 5 | 1 582.353 6 | 1 438.120 4 | 1 371.050 3 | 1 220.404 2 | 1 000.213 6 |
| 泾县 JX | 0.545 8 | 0.000 0 | 237.553 6 | 123.765 5 | 2 042.560 1 | 1 898.927 0 | 1 831.920 9 | 1 683.540 6 | 1 464.721 7 |
| 仙居 XJ | 0.251 9 | 0.674 2 | 0.000 0 | 146.794 0 | 2 251.480 5 | 2 107.132 3 | 2 040.069 9 | 1 888.425 4 | 1 667.240 1 |
| 遂昌 SC | 0.508 1 | 0.390 0 | 0.460 7 | 0.000 0 | 2 104.708 5 | 1 960.372 6 | 1 893.308 7 | 1 741.786 6 | 1 520.714 2 |
| 元谋 YM | 0.480 5 | 0.340 3 | 0.439 2 | 0.071 0 | 0.000 0 | 144.614 4 | 211.546 0 | 367.289 9 | 589.458 9 |
| 宾川 BC | 0.524 9 | 0.364 8 | 0.395 1 | 0.140 2 | 0.074 1 | 0.000 0 | 67.070 2 | 223.452 1 | 445.148 1 |
| 武定 WD | 0.013 4 | 0.514 3 | 0.316 2 | 0.538 1 | 0.513 9 | 0.554 7 | 0.000 0 | 158.489 4 | 378.991 6 |
| 师宗 SZ | 0.046 1 | 0.631 1 | 0.220 6 | 0.501 9 | 0.475 3 | 0.548 4 | 0.092 2 | 0.000 0 | 222.460 2 |
| 册亨 CH | 0.747 4 | 0.748 9 | 0.791 5 | 0.913 6 | 0.896 5 | 0.957 0 | 0.753 2 | 0.731 2 | 0.000 0 |
表8 毛红椿天然居群间的遗传距离(下三角)和地理距离(上三角)
Table 8 Genetic distance (lower triangle) and geographic distance (upper triangle) between natural populations of Toona ciliata var. pubescens
| 居群 Population | 宜丰 YF | 泾县 JX | 仙居 XJ | 遂昌 SC | 元谋 YM | 宾川 BC | 武定 WD | 师宗 SZ | 册亨 CH |
|---|---|---|---|---|---|---|---|---|---|
| 宜丰 YF | 0.000 0 | 466.550 4 | 669.412 6 | 522.624 5 | 1 582.353 6 | 1 438.120 4 | 1 371.050 3 | 1 220.404 2 | 1 000.213 6 |
| 泾县 JX | 0.545 8 | 0.000 0 | 237.553 6 | 123.765 5 | 2 042.560 1 | 1 898.927 0 | 1 831.920 9 | 1 683.540 6 | 1 464.721 7 |
| 仙居 XJ | 0.251 9 | 0.674 2 | 0.000 0 | 146.794 0 | 2 251.480 5 | 2 107.132 3 | 2 040.069 9 | 1 888.425 4 | 1 667.240 1 |
| 遂昌 SC | 0.508 1 | 0.390 0 | 0.460 7 | 0.000 0 | 2 104.708 5 | 1 960.372 6 | 1 893.308 7 | 1 741.786 6 | 1 520.714 2 |
| 元谋 YM | 0.480 5 | 0.340 3 | 0.439 2 | 0.071 0 | 0.000 0 | 144.614 4 | 211.546 0 | 367.289 9 | 589.458 9 |
| 宾川 BC | 0.524 9 | 0.364 8 | 0.395 1 | 0.140 2 | 0.074 1 | 0.000 0 | 67.070 2 | 223.452 1 | 445.148 1 |
| 武定 WD | 0.013 4 | 0.514 3 | 0.316 2 | 0.538 1 | 0.513 9 | 0.554 7 | 0.000 0 | 158.489 4 | 378.991 6 |
| 师宗 SZ | 0.046 1 | 0.631 1 | 0.220 6 | 0.501 9 | 0.475 3 | 0.548 4 | 0.092 2 | 0.000 0 | 222.460 2 |
| 册亨 CH | 0.747 4 | 0.748 9 | 0.791 5 | 0.913 6 | 0.896 5 | 0.957 0 | 0.753 2 | 0.731 2 | 0.000 0 |
| [1] | Baranek M, Kadlec M, Raddová J, Vachůn M, Pidra M (2001). Evalutation of RAPD data by POPGENE 32 software. Proceedings of 9th International Conference of Horticulture, September 3th-6th Lednice, Czech Republic, 2, 465-472. |
| [2] |
Channell R, Lomolino MV (2000). Dynamic biogeography and conservation of endangered species. Nature, 403, 84-86.
DOI URL PMID |
| [3] | Coates DJ (2000). Defining conservation units in a rich and fragmented flora: implications for the management of genetic resources and evolutionary processes in south- west Australian plants. Australian Journal of Botany, 48, 329-339. |
| [4] |
Eckert CG, Samis KE, Lougheed SC (2008). Genetic variation across species’ geographical ranges: the central- marginal hypothesis and beyond. Molecular Ecology, 17, 1170-1188.
DOI URL PMID |
| [5] |
Eckstein RL, O’Neill RA, Danihelka J, Otte A, Köhler W (2006). Genetic structure among and within peripheral and central populations of three endangered floodplain violets. Molecular Ecology, 15, 2367-2379.
DOI URL PMID |
| [6] |
Excoffier L, Lischer HEL (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567.
URL PMID |
| [7] | Gapare WJ, Aitken SN, Ritland CE (2005). Genetic diversity of core and peripheral Sitka spruce (Picea sitchensis(Bong.) Carr) populations: implications for conservation of widespread species. Biological Conservation, 123, 113-123. |
| [8] | Goverde M, Schweier K, Baur B, Erhardt A (2002). Small- scale habitat fragmentation effects on pollinator behaviour: experimental evidence from the bumblebee Bombus veteranus on calcareous grasslands. Biological Conservation, 104, 293-299. |
| [9] | Groom MJ (1977). Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. The American Naturalist, 111, 1169-1194. |
| [10] |
Guries RP, Ledig FT (1982). Genetic diversity and population structure in pitch pine (Pinus rigida Mill.). Evolution, 36, 387-402.
URL PMID |
| [11] | Hamann A, El-Kassaby YA, Koshy MP, Namkoong G (1998). Multivariate analysis of allozymic and quantitative trait variation in (Alnus rubra Bong.): geographic patterns and evolutionary implication. Canadian Journal of Forest Research, 10, 1557-1565. |
| [12] |
Hampe A, Petit RJ (2005). Conserving biodiversity under climate change: the rear edge matters. Ecology Letters, 8, 461-467.
DOI URL PMID |
| [13] | Hamrick JL, Godt MJW, Murawski DA, Loveless MD (1991). Correlation between species traits and allozyme diversity: implication for conservation biology. In: Falk DA, Holsinger KE eds. Genetics and Conservation of Rare Plants. Oxford University Press, New York. |
| [14] |
Huang ZH, Liu NF, Zhou TL (2005). A comparative study of genetic diversity of peripheral and central populations of Chukar partridge from northwestern China. Biochemical Genetics, 43, 613-621.
DOI URL PMID |
| [15] |
Kirkpatrick M, Ravigné V (2002). Speciation by natural and sexual selection: models and experiments. The American Naturalist, 159, S22-S35.
URL PMID |
| [16] | Lammi A, Siikamaki P, Mustajarvi K (1999). Genetic diversity, population size and fitness in central and peripheral populations of a rare plant Lychnis viscaria. Conservation Biology, 13, 1069-1078. |
| [17] | Ledig FT (1986). Conservation strategies for forest gene resources. Forest Ecology and Management, 14, 77-90. |
| [18] | Lesica P, Allendorf FW (1995). When are peripheral populations valuable for conservation? Conservation Biology, 9, 753-760. |
| [19] |
Li L, Sun ZX, Yang SD, Chang LR, Yang LH (2006). Analysis of genetic variation of Abalone (Haliotis discus hannal) populations with microsatellite markers. Hereditas, 28, 1549-1554. (in Chinese with English abstract)
URL PMID |
|
[ 李莉, 孙振兴, 杨树德, 常林瑞, 杨立红 (2006). 用微卫星标记分析皱纹盘鲍群体的遗传变异. 遗传, 28, 1549-1554.]
PMID |
|
| [20] | Liu J, Chen YT, Sun ZX, Jiang JM, He GP, Rao LB, Wu TL (2008). Spatial genetic structure of Toona ciliata var. pubescens populations in terms of spatial autocorrelation analysis. Scientia Silvae Sinicae, 44(6), 53-59. (in Chinese with English abstract) |
| [ 刘军, 陈益泰, 孙宗修, 姜景民, 何贵平, 饶龙兵, 吴天林 (2008). 基于空间自相关分析研究毛红椿天然居群的空间遗传结构. 林业科学, 44(6), 53-59.] | |
| [21] | Liu J, Sun ZX, Chen YT, He GP, Wu TL (2006). Isolation of microsatellite DNA from endangered tree species Toona ciliata var. pubescens and optimization of SSR reaction system. China Biotechnology, 26(12), 50-55. (in Chinese with English abstract) |
| [ 刘军, 孙宗修, 陈益泰, 何贵平, 吴天林 (2006). 珍稀濒危树种毛红椿微卫星DNA分离及SSR反应体系优化. 中国生物工程杂志, 26(12), 50-55.] | |
| [22] | Marshall DR, Brown ADH (1975). Optimum sampling strategies for gene conservation. In: Frankel OH, Hawkes JG eds. Crop Genetic Resources for Today and Tomorrow. Cambridge University Press, Cambridge. 53-80. |
| [23] | Matsumura C, Washitani I (2000). Effects of population size and pollinator limitation on seed-set of Primula sieboldii populations in a fragmented landscape. Ecological Research, 15, 307-322. |
| [24] | Mayr E (1963). Animal Species and Evolution. Harvard University Press, Cambridge. |
| [25] | Miller MP (1997). Tools for Population Genetic Analysis(TFPGA) version 1.3. Department of Biological Sciences, Northern Arizona University, Flagstaff. |
| [26] |
Nei M, Maruyama T, Chakrorty R (1975). The bottleneck effect and genetic variability in populations. Evolution, 29, 1-10.
DOI URL PMID |
| [27] | Peakall R, Smouse PE (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6, 288-295. |
| [28] |
Takezakin N, Nei M (1996). Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics, 144, 389-399.
URL PMID |
| [29] |
Thomas CD, Bodsworth EJ, Wilson RJ, Simmons AD, Davies ZG, Musche M, Conradt L (2001). Ecological and evolutionary processes at expanding range margins. Nature, 411, 577-581.
DOI URL PMID |
| [30] | van Rossum F, Vekemans X, Gratia E, Meerts P (2003). A comparative study of allozyme variation of peripheral and central populations of Silene nutans L.(Caryophyllaceae) from Western Europe: implications for conservation. Plant Systematics and Evolution, 242, 49-61. |
| [31] | Wang YQ, Zhang ZB, Xu LX (2002). The genetic diversity of central and peripheral populations of ratlike hamster (Cricetulus triton). Chinese Science Bulletin, 47, 201-206. |
| [32] | White TL, Adams WT, Neale DB (2007). Forest Genetics. CABI Publishing, New York. 259-284. |
| [33] | Wright S (1965). The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution, 19, 395-420. |
| [1] | 陈天翌, 娄安如. 青藏高原东侧白桦种群的遗传多样性与遗传结构[J]. 植物生态学报, 2022, 46(5): 561-568. |
| [2] | 张新新, 王茜, 胡颖, 周玮, 陈晓阳, 胡新生. 植物边缘种群遗传多样性研究进展[J]. 植物生态学报, 2019, 43(5): 383-395. |
| [3] | 张俪文, 韩广轩. 植物遗传多样性与生态系统功能关系的研究进展[J]. 植物生态学报, 2018, 42(10): 977-989. |
| [4] | 王锦楠, 陈进福, 陈武生, 周新洋, 许东, 李际红, 亓晓. 柴达木地区野生黑果枸杞种群遗传多样性的AFLP分析[J]. 植物生态学报, 2015, 39(10): 1003-1011. |
| [5] | 张炜, 罗建勋, 辜云杰, 胡庭兴. 西南地区麻疯树天然种群遗传多样性的等位酶变异[J]. 植物生态学报, 2011, 35(3): 330-336. |
| [6] | 魏源, 王世杰, 刘秀明, 黄天志. 不同喀斯特小生境中土壤丛枝菌根真菌的遗传多样性[J]. 植物生态学报, 2011, 35(10): 1083-1090. |
| [7] | 张云红, 侯艳, 娄安如. 华北地区小丛红景天种群的AFLP遗传多样性[J]. 植物生态学报, 2010, 34(9): 1084-1094. |
| [8] | 刘伟, 王曦, 干友民, 黄林凯, 谢文刚, 苗佳敏. 高山嵩草种群在放牧干扰下遗传多样性的变化[J]. 植物生态学报, 2009, 33(5): 966-973. |
| [9] | 陈良华, 胡庭兴, 张帆, 李国和. 用AFLP技术分析四川核桃资源的遗传多样性[J]. 植物生态学报, 2008, 32(6): 1362-1372. |
| [10] | 周会平, 陈进, 张寿洲. 具混合繁殖策略的草本植物异果舞花姜的居群遗传结构[J]. 植物生态学报, 2008, 32(4): 751-759. |
| [11] | 严茂粉, 李向华, 王克晶. 北京地区野生大豆种群SSR标记的遗传多样性评价[J]. 植物生态学报, 2008, 32(4): 938-950. |
| [12] | 刘亚令, 李作洲, 姜正旺, 刘义飞, 黄宏文. 中华猕猴桃和美味猕猴桃自然居群遗传结构及其种间杂交渐渗[J]. 植物生态学报, 2008, 32(3): 704-718. |
| [13] | 魏宇昆, 高玉葆. 禾草内生真菌的遗传多样性及其共生关系[J]. 植物生态学报, 2008, 32(2): 512-520. |
| [14] | 杨明博, 杨劼, 杨九艳, 梁娜, 清华. 鄂尔多斯高原不同生境条件下中间锦鸡儿植物叶片表皮特征及遗传多样性变化分析[J]. 植物生态学报, 2007, 31(6): 1181-1189. |
| [15] | 穆立蔷, 刘赢男. 不同地理分布区紫椴种群的遗传多样性变化[J]. 植物生态学报, 2007, 31(6): 1190-1198. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
Copyright © 2022 版权所有 《植物生态学报》编辑部
地址: 北京香山南辛村20号, 邮编: 100093
Tel.: 010-62836134, 62836138; Fax: 010-82599431; E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
备案号: 京ICP备16067583号-19