

植物生态学报 ›› 2011, Vol. 35 ›› Issue (5): 531-538.DOI: 10.3724/SP.J.1258.2011.00531
所属专题: 生物多样性
张利锐, 彭艳玲, 任广朋, 周永锋, 李忠虎, 刘建全*( )
)
收稿日期:2010-08-03
接受日期:2010-12-13
出版日期:2011-08-03
发布日期:2011-06-07
通讯作者:
刘建全
作者简介:* E-mail: liujq@nnwipb.ac.cn
ZHANG Li-Rui, PENG Yan-Ling, REN Guang-Peng, ZHOU Yong-Feng, LI Zhong-Hu, LIU Jian-Quan*( )
)
Received:2010-08-03
Accepted:2010-12-13
Online:2011-08-03
Published:2011-06-07
Contact:
LIU Jian-Quan
摘要:
利用两个核基因座位C3H和GI, 对重叠分布于中国东南部的两个松属(Pinus)物种马尾松(P. massoniana)和黄山松(P. hwangshanensis)的22个群体88个个体进行了遗传多样性和种间分化模式研究。在这两个核基因座位上, 两种植物都表现出较低的核苷酸多样性水平(马尾松πsil = 0.001 71; 黄山松πsil = 0.003 40), 但是马尾松要显著低于黄山松; 在种内分化水平上, 马尾松的种内遗传分化也明显低于黄山松(马尾松FST = 0.059; 黄山松FST = 0.339)。这可能是由于黄山松的海拔分布高于马尾松, 而高海拔分布使黄山松的分布区域更加片段化, 促使其形成较高的种内遗传多样性和遗传分化。分子变异分析(AMOVA)发现, 两物种基于两个核基因座位的种间差异为48.86%, 而GI基因座位上的种间差异明显高于C3H座位(GI: 77.24%, C3H: 20.48%), 同时, 基因谱系显示两物种的共享单倍型仅在C3H座位上存在。结合这两个基因的功能, 推测GI基因可能在物种形成过程中受到了一定的选择压力, 因为GI基因参与调控植物的开花时间, 而C3H与木质素表达水平的调控有关。不同的选择压力使得GI的进化速度相对较快, 从而加速了黄山松和马尾松的物种分化。
张利锐, 彭艳玲, 任广朋, 周永锋, 李忠虎, 刘建全. 马尾松和黄山松两个核基因位点的群体遗传多样性和种间分化. 植物生态学报, 2011, 35(5): 531-538. DOI: 10.3724/SP.J.1258.2011.00531
ZHANG Li-Rui, PENG Yan-Ling, REN Guang-Peng, ZHOU Yong-Feng, LI Zhong-Hu, LIU Jian-Quan. Population genetic diversity and species divergence of Pinus massoniana and P. hwangshanensis at two nucleotide loci. Chinese Journal of Plant Ecology, 2011, 35(5): 531-538. DOI: 10.3724/SP.J.1258.2011.00531
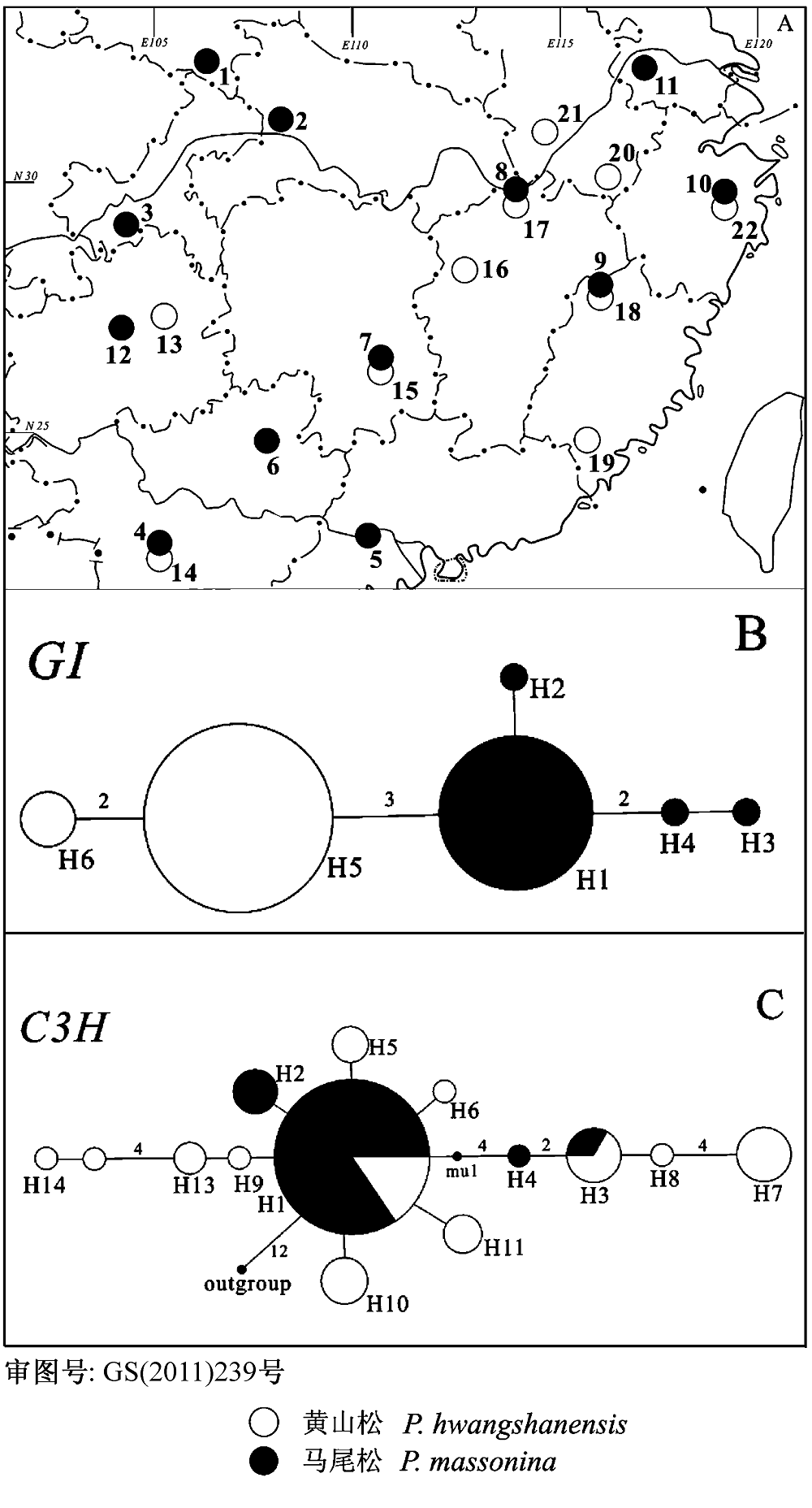
图1 马尾松和黄山松群体采样地点分布(A)和核基因座位C3H和GI的基因谱系(B, C)。
Fig. 1 Sampling localities of Pinus massoniana and P. hwangshanensis (A) and gene genealogies of nuclear loci C3H and GI (B, C).
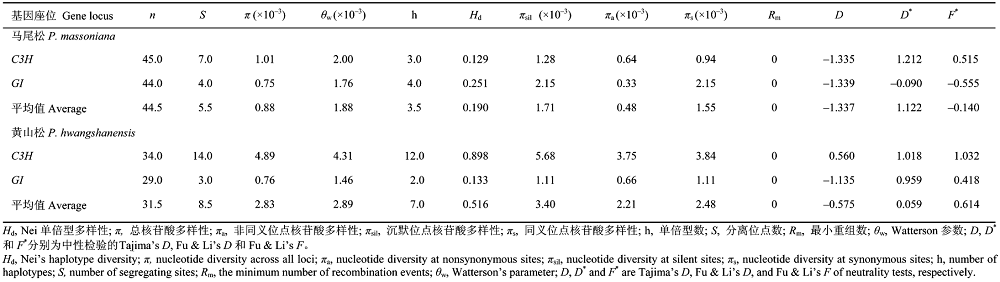 |
表2 马尾松和黄山松核基因座位C3H和GI核苷酸多样性和中性检测
Table 2 Nucleotide diversity and neutrality tests over the C3H and GI locus in Pinus massoniana and P. hwangshanensis
 |
| 变异来源 Source of variation (%) | 遗传分化系数 Coefficient of gene differentiation (FST) | ||||
|---|---|---|---|---|---|
| 基因座位 Gene locus | 种间 Among species | 种内群体间 Among populations within species | 群体内 Within population | 马尾松 P. massoniana | 黄山松 P. hwangshanensis |
| C3H | 20.48 | 15.81 | 63.71 | 0.071 | 0.210* |
| GI | 77.24 | 6.99 | 15.77 | 0.047 | 0.467** |
| 平均值 Average | 48.86 | 11.35 | 39.74 | 0.059 | 0.339 |
表3 马尾松和黄山松核基因座位C3H和GI变异的分子方差(AMOVA)分析(10 000次重复)
Table 3 Analysis of molecular variance (AMOVA) of nuclear loci C3H and GI variation in Pinus massoniana and P. hwangshanensis (10 000 permutations)
| 变异来源 Source of variation (%) | 遗传分化系数 Coefficient of gene differentiation (FST) | ||||
|---|---|---|---|---|---|
| 基因座位 Gene locus | 种间 Among species | 种内群体间 Among populations within species | 群体内 Within population | 马尾松 P. massoniana | 黄山松 P. hwangshanensis |
| C3H | 20.48 | 15.81 | 63.71 | 0.071 | 0.210* |
| GI | 77.24 | 6.99 | 15.77 | 0.047 | 0.467** |
| 平均值 Average | 48.86 | 11.35 | 39.74 | 0.059 | 0.339 |
| [1] |
Abbott RJ, Comes HP (2004). Evolution in the Arctic: a phylogeographic analysis of the circumarctic plant, Saxifraga oppositifolia (purple saxifrage). New Phytologist, 161, 211-224.
DOI URL |
| [2] | Akey JM, Eberle MA, Rieder MJ, Carlson CS, Shriver MD, Nickerson DA, Kruglyak L (2004). Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biology, 2, 1591-1599. |
| [3] |
Arunyawat U, Stephan W, Städler T (2007). Using multilocus sequence data to assess population structure, natural selection, and linkage disequilibrium in wild tomatoes. Molecular Biology and Evolution, 24, 2310-2322.
URL PMID |
| [4] |
Bandelt HJ, Forster P, Röhl A (1999). Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37-48.
DOI URL PMID |
| [5] |
Belahbib N, Pemonge MH, Ouassou A, Sbay H, Kremer A, Petit RJ (2001). Frequent cytoplasmic exchanges between oak species that are not closely related: Quercus suber and Q. ilex in Morocco. Molecular Ecology, 10, 2003-2012.
URL PMID |
| [6] |
Bomblies K, Weigel D (2007). Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nature Reviews Genetics, 8, 382-393.
URL PMID |
| [7] |
Bouillé M, Bousquet J (2005). Trans-species shared polymorphism at orthologous nuclear gene loci among distant species in the conifer Picea (Pinaceae): implications for the long-term maintenance of genetic diversity in trees. American Journal of Botany, 92, 63-73.
DOI URL PMID |
| [8] |
Brown GR, Gill GP, Kuntz RJ, Langley CH, Neale DB (2004). Nucleotide diversity and linkage disequilibrium in Loblolly pine. Proceedings of the National Academy of Sciences of the United Stated of America, 101, 15255-15260.
DOI URL |
| [9] |
Chase CD (2007). Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends in Genetics, 23, 81-90.
URL PMID |
| [10] |
Chen J, Källman T, Gyllenstrand N, Lascoux M (2010). New insights on the speciation history and nucleotide diversity of three boreal spruce species and a Tertiary relict. Heredity, 104, 3-14.
URL PMID |
| [11] |
Chiang YC, Hung KH, Schaal BA, Ge XJ, Tsu TW, Chiang TY (2006). Contrasting phylogeographical patterns between mainland and island taxa of the Pinus luchuensis complex. Molecular Ecology, 15, 765-779.
URL PMID |
| [12] | Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19, 11-15. |
| [13] |
Dumolin-Lapègue S, Pemonge MH, Petit RJ (1997). An enlarged set of consensus primers for the study of organelle DNA in plants. Molecular Ecology, 6, 393-397.
DOI URL PMID |
| [14] |
Excoffier L, Laval G, Schneider S (2005). Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online, 1, 47-50.
URL PMID |
| [15] |
Fu YX, Li WH (1993). Statistical tests of neutrality of mutations. Genetics, 133, 693-709.
URL PMID |
| [16] | Fu LG, Li N, Elias TS, Mill RR (1999). Pinaceae. In: Wu ZY, Raven PH eds. Flora of China, Volume 4. Science Press, Beijing. 17. |
| [17] |
García-Gil MR, Mikkonen M, Savolainen O (2003). Nucleotide diversity at two phytochrome loci along a latitudinal cline in Pinus sylvestris. Molecular Ecology, 12, 1195-1206.
DOI URL PMID |
| [18] |
Heuertz M, de Paoli E, Källman T, Larsson H, Jurman I, Morgante M, Lascoux M, Gyllenstrand N (2006). Multilocus patterns of nucleotide diversity, linkage disequilibrium and demographic history of Norway spruce (Picea abies (L.) Karst). Genetics, 174, 2095-2105.
URL PMID |
| [19] |
Hoballah ME, Gubitz T, Stuurman J, Broger L, Barone M, Mandel T, Dell’Olivo A, Arnold ML, Kuhlemeier C (2007). Single gene-mediated shift in pollinator attraction in Petunia. The Plant Cell, 19, 779-790.
DOI URL PMID |
| [20] |
Huq E, Tepperman JM, Quail PH (2000). GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proceedings of the National Academy of Sciences of the United Stated of America, 97, 9789-9794.
DOI URL |
| [21] | Hu XS (胡新生) (2002). A review on understanding the genetic structure of population. Scientia Silvae Sinicae (林业科学), 38, 119-128. (in Chinese with English abstract) |
| [22] |
Hudson RR, Kaplan NL (1985). Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics, 111, 147-164.
URL PMID |
| [23] |
Kado T, Yoshimaru H, Tsumura Y, Tachida H (2003). DNA variation in a conifer, Cryptomeria japonica (Cupressa- ceae sensu lato). Genetics, 164, 1547-1599.
URL PMID |
| [24] |
Kumar S, Tamura K, Nei M (2004). MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics, 5, 150-163.
DOI URL PMID |
| [25] |
Ledig FT (2000). Founder effects and the genetic structure of Coulter pine. Journal of Heredity, 91, 307-315.
URL PMID |
| [26] |
Li Y, Stocks M, Hemmilä S, Källman T, Zhu HT, Zhou YF, Chen J, Liu JQ, Lascoux M (2010a). Demographic histories of four spruce (Picea) species of the Qinghai-Tibetan Plateau and neighboring areas inferred from multiple nuclear loci. Molecular Biology and Evolution, 27, 1001-1014.
DOI URL PMID |
| [27] |
Li SX, Chen Y, Gao HD, Yin TM (2010b). Potential chromosomal introgression barriers revealed by linkage analysis in a hybrid of Pinus massoniana and P. hwangshanensis. BMC Plant Biology, 10, doi: 10.1186/1471-2229-10-37.
DOI URL PMID |
| [28] |
Li ZH, Zhang Q, Liu JQ, Källman T, Lascoux M (2011). The Pleistocene demography of an alpine juniper of the Qinghai-Tibetan Plateau: tabula rasa, cryptic refugia or something else? Journal of Biogeography, 38, 31-43.
DOI URL |
| [29] | Luo SJ (罗世家), Zou HY (邹惠渝), Liang SW (梁师文) (2001). Study on the introgressive hybridization between Pinus hwangshanensis and P. massoniana. Scientia Silvae Sinicae (林业科学), 6, 118-122. (in Chinese with English abstract) |
| [30] |
Ma XF, Szmidt AE, Wang XR (2006). Genetic structure and evolutionary history of a diploid hybrid pine Pinus densata inferred from the nucleotide variation at seven gene loci. Molecular Biology and Evolution, 23, 807-816.
DOI URL PMID |
| [31] |
Mallet J (2007). Hybrid speciation. Nature, 446, 279-283.
URL PMID |
| [32] | Mayr E (1942). Systematics and the Origin of Species. Columbia University Press, New York, USA. |
| [33] | Nei M (1987). Molecular Evolutionary Genetics. Columbia University Press, New York, USA. |
| [34] |
Orr HA, Masly JP, Presgraves DC (2004). Speciation genes. Current Opinion in Genetics and Development, 14, 675-679.
DOI URL PMID |
| [35] |
Ralph J, Akiyama T, Kim H, Lu F, Schatz PF, Marita JM, Ralph SA, Reddy MSS, Chen F, Dixon R (2006). Effects of coumarate 3-hydroxylase down-regulation on lignin structure. The Journal of Biological Chemistry, 281, 8843-8853.
URL PMID |
| [36] |
Ren GP, Abbott R, Zhou YF, Zhang LR, Peng YL, Liu JQ (2010). Genetic divergence, range expansion and homoploid hybrid speciation among four pine species in northeast China. Molecular Ecology. (under review)
DOI URL PMID |
| [37] |
Rieseberg LH, Kim SC, Randell RA, Whitney KD, Gross BL, Lexer C, Clay K (2007). Hybridization and the colonization of novel habitats by annual sunflowers. Genetica, 129, 149-165.
DOI URL |
| [38] |
Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics, 19, 2496-2497.
DOI URL PMID |
| [39] |
Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA (1998). Phylogeographic studies in plants: problems and prospects. Molecular Ecology, 7, 465-474.
DOI URL |
| [40] |
Strasburg JL, Rieseberg LH (2008). Molecular demographic history of the annual sunflowers Helianthus annuus and H. petiolaris—large effective population sizes and rates of long-term gene flow. Evolution, 62, 1936-1950.
DOI URL PMID |
| [41] |
Tajima F (1983). Evolutionary relationship of DNA sequences in finite populations. Genetics, 105, 437-460.
URL PMID |
| [42] |
Tajima F (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123, 585-595.
URL PMID |
| [43] |
Wakeley J (2001). The coalescent in an island model of population subdivision with variation among demes. Theoretical Population Biology, 59, 133-144.
DOI URL PMID |
| [44] |
Wang HW, Ge S (2006). Phylogeography of the endangered Cathaya argyrophylla (Pinaceae) inferred from sequence variation of mitochondrial and nuclear DNA. Molecular Ecology, 15, 4109-4122.
DOI URL PMID |
| [45] |
Watterson GA (1975). On the number of segregating sites in genetical models without recombination. Theoretical Population Biology, 7, 256-276.
DOI URL PMID |
| [46] |
Willyard A, Cronn R, Liston A (2009). Reticulate evolution and incomplete lineage sorting among the Ponderosa pines. Molecular Phylogenetics and Evolution, 52, 498-511.
DOI URL PMID |
| [47] |
Wright S (1951). The genetical structure of populations. Annals of Eugenics, 15, 323-354.
DOI URL PMID |
| [48] |
Wright SI, Gaut BS (2005). Molecular population genetics and the search for adaptive evolution in plants. Molecular Biology and Evolution, 22, 506-519.
DOI URL PMID |
| [49] |
Wu CI (2001). The genic view of the process of speciation. Journal of Evolutionary Biology, 14, 851-865.
DOI URL |
| [50] | Xing YH (邢有华), Fang YX (方永鑫), Wu GR (吴根荣) (1992). The preliminary study of natural hybridizations between Pinus hwangshanensis and P. massoniana in Dabie Mountain of Anhui Province. Anhui Forestry Science and Technology (安徽林业科技), 4, 5-9. (in Chinese) |
| [51] |
Zheng XM, Ge S (2010). Ecological divergence in the presence of gene flow in two closely related Oryza species (Oryza rufipogon and O. nivara). Molecular Ecology, 19, 2439-2454.
DOI URL PMID |
| [52] |
Zhou YF, Abbott RJ, Jiang ZY, Du FK, Milne RI, Liu JQ (2010). Gene flow and species delimitation: a case study of two pine species with overlapping distributions in southeast China. Evolution, 64, 2342-2352.
DOI URL PMID |
| [53] | Zou YH (邹惠渝), Li Z (黎志), Luo SJ (罗世家), Yi YM (易咏梅), Wang JR (王静茹) (2003). Study on taxonomy of Pinus hwangshanensis. Bulletin of Botanical Research (植物研究), 23, 278-279. (in Chinese with English abstract) |
| [1] | 陈雨婷, 马松梅, 张丹, 张林, 王春成. 新疆同域分布梭梭和白梭梭多样性格局及其形成机制[J]. 植物生态学报, 2024, 48(1): 56-67. |
| [2] | 杨鑫, 任明迅. 环南海区域红树物种多样性分布格局及其形成机制[J]. 植物生态学报, 2023, 47(8): 1105-1115. |
| [3] | 郑炀, 孙学广, 熊洋阳, 袁贵云, 丁贵杰. 叶际微生物对马尾松凋落针叶分解的影响[J]. 植物生态学报, 2023, 47(5): 687-698. |
| [4] | 王春成, 张云玲, 马松梅, 黄刚, 张丹, 闫涵. 中国扁桃亚属四种野生扁桃的系统发育与物种分化[J]. 植物生态学报, 2021, 45(9): 987-995. |
| [5] | 罗斯生, 罗碧珍, 魏书精, 胡海清, 李小川, 吴泽鹏, 王振师, 周宇飞, 钟映霞. 中度强度森林火灾对马尾松次生林土壤有机碳密度的影响[J]. 植物生态学报, 2020, 44(10): 1073-1086. |
| [6] | 宋平, 张蕊, 周志春, 童建设, 王晖. 局部供氮对低磷胁迫下马尾松不同家系生长及根系参数的影响[J]. 植物生态学报, 2017, 41(6): 622-631. |
| [7] | 宋思梦, 张丹桔, 张健, 杨万勤, 张艳, 周扬, 李勋. 马尾松人工林林窗边缘效应对油樟化学计量特征的影响[J]. 植物生态学报, 2017, 41(10): 1081-1090. |
| [8] | 姜超, 谭珂, 任明迅. 季风对亚洲热带植物分布格局的影响[J]. 植物生态学报, 2017, 41(10): 1103-1112. |
| [9] | 汪沁, 杨万勤, 吴福忠, 张健, 谭波, 张玺涛. 马尾松人工林伐桩储量与分解特征[J]. 植物生态学报, 2016, 40(5): 458-468. |
| [10] | 宋平, 张蕊, 张一, 周志春, 丰忠平. 模拟氮沉降对低磷胁迫下马尾松无性系细根形态和氮磷效率的影响[J]. 植物生态学报, 2016, 40(11): 1136-1144. |
| [11] | 张艳, 张丹桔, 张健, 杨万勤, 邓长春, 李建平, 李勋, 唐仕姗, 张明锦. 马尾松人工林林窗大小对两种凋落叶难降解物质含量的影响[J]. 植物生态学报, 2015, 39(8): 785-796. |
| [12] | 陈云玉, 熊德成, 黄锦学, 王韦韦, 胡双成, 邓飞, 许辰森, 冯建新, 史顺增, 钟波元, 陈光水. 中亚热带不同演替阶段的马尾松和米槠人工林的细根生产量研究[J]. 植物生态学报, 2015, 39(11): 1071-1081. |
| [13] | 崔宁洁,张丹桔,刘洋,张健,杨万勤,欧江,张捷,宋小艳,殷睿. 马尾松人工林不同大小林窗植物多样性及其季节动态[J]. 植物生态学报, 2014, 38(5): 477-490. |
| [14] | 庞丽, 张一, 周志春, 丰忠平, 储德裕. 模拟氮沉降对低磷胁迫下马尾松不同家系根系分泌和磷效率的影响[J]. 植物生态学报, 2014, 38(1): 27-35. |
| [15] | 谭小梅, 周志春, 金国庆, 张一. 马尾松二代无性系种子园子代父本分析及花粉散布[J]. 植物生态学报, 2011, 35(9): 937-945. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
Copyright © 2022 版权所有 《植物生态学报》编辑部
地址: 北京香山南辛村20号, 邮编: 100093
Tel.: 010-62836134, 62836138; Fax: 010-82599431; E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
备案号: 京ICP备16067583号-19