

植物生态学报 ›› 2025, Vol. 49 ›› Issue (10): 1677-1684.DOI: 10.17521/cjpe.2025.0008 cstr: 32100.14.cjpe.2025.0008
所属专题: 濒危植物种群特征与保护
黄承玲1, 黎荣瀚1, 覃红玲2, 杨胜雄2, 田晓玲2, 夏国威1, 陈正仁3, 周玮1,*( )
)
收稿日期:2025-01-07
接受日期:2025-05-01
出版日期:2025-10-20
发布日期:2025-11-20
通讯作者:
*周玮(605466767@qq.com)基金资助:
HUANG Cheng-Ling1, LI Rong-Han1, QIN Hong-Ling2, YANG Sheng-Xiong2, TIAN Xiao-Ling2, XIA Guo-Wei1, CHEN Zheng-Ren3, ZHOU Wei1,*( )
)
Received:2025-01-07
Accepted:2025-05-01
Online:2025-10-20
Published:2025-11-20
Supported by:摘要: 荔波杜鹃(Rhododendron liboense)是分布于喀斯特石灰岩山顶的极小种群植物, 具有较高的观赏和科学研究价值, 其生境脆弱且野生种群稀少, 亟待开展保护遗传学研究。该研究以荔波杜鹃3个种群的43株个体为对象, 采用简化基因组测序技术(ddRAD-seq)获得单核苷酸多态性(SNP)数据, 分析荔波杜鹃种群的遗传多样性和遗传结构, 并推断种群历史动态。结果表明: 荔波杜鹃的遗传多样性相对较低(期望杂合度(He) = 0.139 62 ± 0.003 32, 核苷酸多样性(π) = 0.157 64 ± 0.003 83), 种群间遗传分化水平中等(遗传分化系数(FST) = 0.075 8), 种群内变异(88.22%)大于种群间变异(11.78%)。Structure分析、主成分分析和聚类分析将荔波杜鹃的3个种群划分为2个遗传分组。种群历史动态分析表明, 荔波杜鹃的有效种群大小从末次冰消期后开始持续上升, 可能与气候逐步变得温暖适宜其生存有关。基于上述研究结果和种群现状, 建议将荔波杜鹃划分两个管理单元进行就地保护, 同时加强人工繁育技术研究, 辅助荔波杜鹃迁地保护和野外回归。
黄承玲, 黎荣瀚, 覃红玲, 杨胜雄, 田晓玲, 夏国威, 陈正仁, 周玮. 基于SNP分子标记的极小种群植物荔波杜鹃保护遗传学. 植物生态学报, 2025, 49(10): 1677-1684. DOI: 10.17521/cjpe.2025.0008
HUANG Cheng-Ling, LI Rong-Han, QIN Hong-Ling, YANG Sheng-Xiong, TIAN Xiao-Ling, XIA Guo-Wei, CHEN Zheng-Ren, ZHOU Wei. Conservation genetics of Rhododendron liboense based on SNP molecular markers, a plant species with extremely small populations. Chinese Journal of Plant Ecology, 2025, 49(10): 1677-1684. DOI: 10.17521/cjpe.2025.0008
| 采样地点 Sampling location | 种群 Population | 采样数 Number of samples | 经度 Longitude | 纬度 Latitude | 海拔 Altitude (m) | 是否在保护区内 Located within the Maolan Natural Reserv (Y/N) |
|---|---|---|---|---|---|---|
| 洞泊 Dongbo | DB | 19 | 107.98° E | 25.37° N | 890 | N |
| 洞塘 Dongtang | DT | 19 | 107.88° E | 25.25° N | 650 | Y |
| 西竹 Xizhu | XZ | 5 | 107.95° E | 25.33° N | 900 | N |
表1 荔波杜鹃采样信息
Table 1 Information on the sampling of Rhododendron liboense
| 采样地点 Sampling location | 种群 Population | 采样数 Number of samples | 经度 Longitude | 纬度 Latitude | 海拔 Altitude (m) | 是否在保护区内 Located within the Maolan Natural Reserv (Y/N) |
|---|---|---|---|---|---|---|
| 洞泊 Dongbo | DB | 19 | 107.98° E | 25.37° N | 890 | N |
| 洞塘 Dongtang | DT | 19 | 107.88° E | 25.25° N | 650 | Y |
| 西竹 Xizhu | XZ | 5 | 107.95° E | 25.33° N | 900 | N |
| 种群 Population | 特有等位基因 Private allele | 期望杂合度 He | 核苷酸多样性 π | 近交系数 FIS |
|---|---|---|---|---|
| DB | 1 076 | 0.149 24 ± 0.002 96 | 0.162 41 ± 0.005 17 | 0.138 26 ± 0.012 71 |
| DT | 1 010 | 0.143 97 ± 0.003 01 | 0.158 15 ± 0.003 14 | 0.307 79 ± 0.038 75 |
| XZ | 357 | 0.125 67 ± 0.003 98 | 0.152 35 ± 0.003 19 | 0.341 78 ± 0.034 49 |
| 平均值 Average value | 814 | 0.139 62 ± 0.003 32 | 0.157 64 ± 0.003 83 | 0.262 61 ± 0.028 65 |
表2 荔波杜鹃种群遗传多样性参数(平均值±标准差)
Table 2 Genetic diversity parameters of Rhododendron liboense populations (mean ± SD)
| 种群 Population | 特有等位基因 Private allele | 期望杂合度 He | 核苷酸多样性 π | 近交系数 FIS |
|---|---|---|---|---|
| DB | 1 076 | 0.149 24 ± 0.002 96 | 0.162 41 ± 0.005 17 | 0.138 26 ± 0.012 71 |
| DT | 1 010 | 0.143 97 ± 0.003 01 | 0.158 15 ± 0.003 14 | 0.307 79 ± 0.038 75 |
| XZ | 357 | 0.125 67 ± 0.003 98 | 0.152 35 ± 0.003 19 | 0.341 78 ± 0.034 49 |
| 平均值 Average value | 814 | 0.139 62 ± 0.003 32 | 0.157 64 ± 0.003 83 | 0.262 61 ± 0.028 65 |
| 变异来源 Source of variation | 自由度 df | 总方差 Sum of squares | 变异组分 Variance components | 变异比 Percentage variation (%) |
|---|---|---|---|---|
| 种群间 Among populations | 2 | 463.938 | 6.910 1 | 11.78 |
| 种群内 Within populations | 81 | 4 169.336 | 51.726 3 | 88.22 |
| 共计 Total | 83 | 4 633.274 | 58.636 4 |
表3 荔波杜鹃种群的分子方差分析(AMOVA)
Table 3 Analysis of molecular variance (AMOVA ) of Rhododendron liboense populations
| 变异来源 Source of variation | 自由度 df | 总方差 Sum of squares | 变异组分 Variance components | 变异比 Percentage variation (%) |
|---|---|---|---|---|
| 种群间 Among populations | 2 | 463.938 | 6.910 1 | 11.78 |
| 种群内 Within populations | 81 | 4 169.336 | 51.726 3 | 88.22 |
| 共计 Total | 83 | 4 633.274 | 58.636 4 |
| 种群 Population | DB | DT | XZ |
|---|---|---|---|
| DB | 0.0894 | 0.0524 | |
| DT | 16.38 | 0.0857 | |
| XZ | 5.24 | 12.73 |
表4 荔波杜鹃群体遗传分化系数(FST)与地理距离
Table 4 Genetic differentiation coefficient (FST) values and geographic distances among Rhododendron liboense populations
| 种群 Population | DB | DT | XZ |
|---|---|---|---|
| DB | 0.0894 | 0.0524 | |
| DT | 16.38 | 0.0857 | |
| XZ | 5.24 | 12.73 |
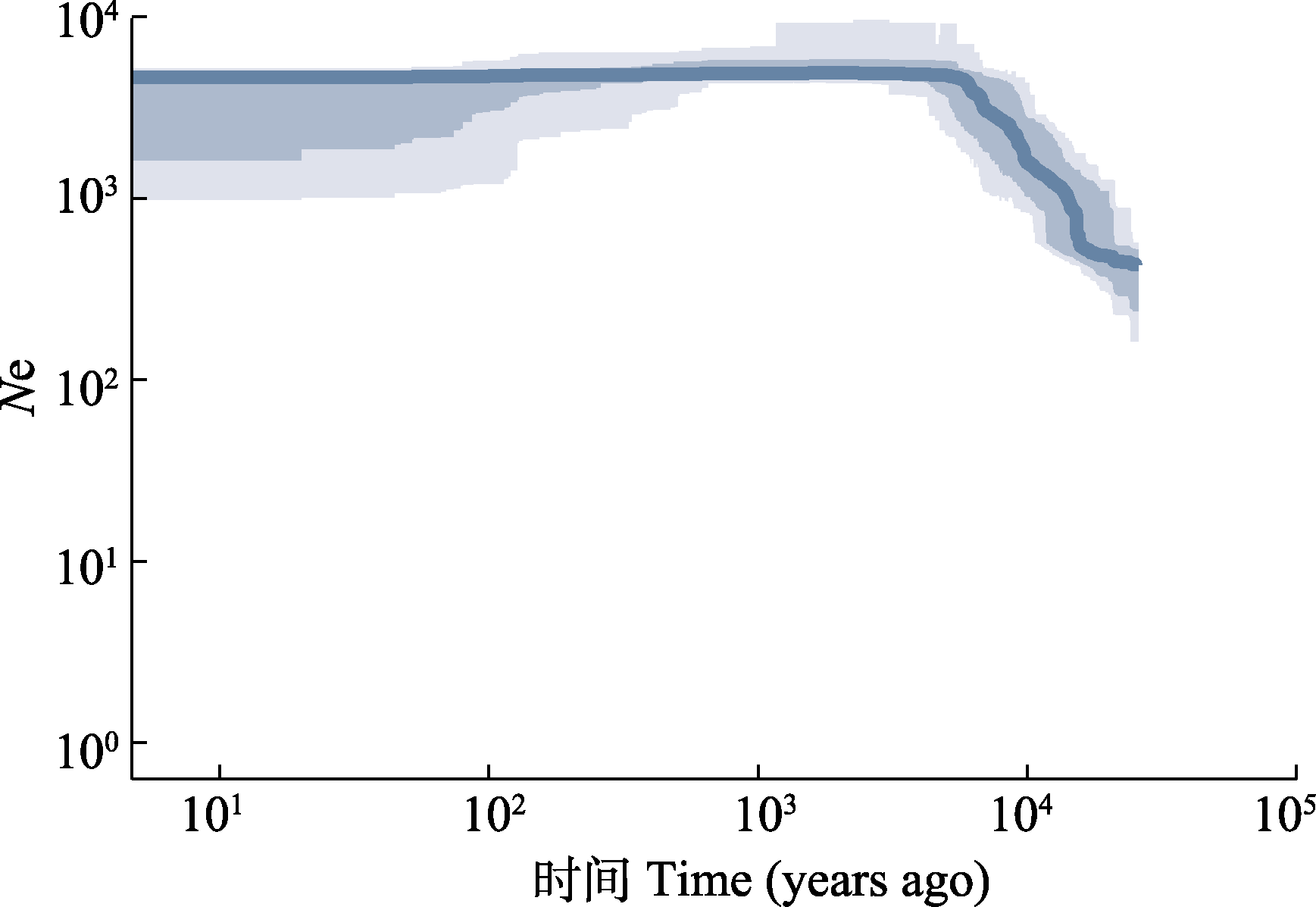
图5 荔波杜鹃种群动态历史分析。浅色和深色阴影分别表示95%和75%置信区间。
Fig. 5 Demographic history of Rhododendron liboense lineages. The light and dark shaded areas represent the 95% and 75% confidence intervals, respectively.
| [1] | Anderson CJ, Tay WT, McGaughran A, Gordon K, Walsh TK (2016). Population structure and gene flow in the global pest, Helicoverpa armigera. Molecular Ecology, 25, 5296-5311. |
| [2] |
Andrews KR, Good JM, Miller MR, Luikart G, Hohenlohe PA (2016). Harnessing the power of RADseq for ecological and evolutionary genomics. Nature Reviews Genetics, 17, 81-92.
DOI PMID |
| [3] | Cao YR (2022). Conservation Biology of Rhododendron hemsleyanum, a Plant Species with Extremely Small Populations. Master degree dissertation, Chinese Academy of Forestry, Beijing. |
| [曹毓蓉 (2022). 极小种群野生植物波叶杜鹃的保护生物学研究. 硕士学位论文, 中国林业科学研究院, 北京.] | |
| [4] | Cao YR, Ma YP, Zhang XJ, Liu XF, Liu DT, Zhang Y, Li ZH, Ma H (2022). Genetic characteristics of Rhododendron Hemsleyanum based on SNP molecular markers. Journal of Yunnan University (Natural Sciences Edition), 44, 859-869. |
| [曹毓蓉, 马永鹏, 张秀姣, 刘雄芳, 刘德团, 张垚, 李正红, 马宏 (2022). 基于SNP分子标记的波叶杜鹃遗传特征分析. 云南大学学报(自然科学版), 44, 859-869.] | |
| [5] | Catchen JM, Hohenlohe PA, Bassham S, Amores A, Cresko WA (2013). Stacks: an analysis tool set for population genomics. Molecular Ecology, 11, 3124-3140. |
| [6] | Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience, 4, 7. DOI: 10.1186/s13742-015-0047-8. |
| [7] | Chen ZR, Lan KM (2003). Rhododendron liboense, a new species of the Ericaceae from Guizhou, China. Acta Phytotaxonomica Sinica, 41, 563-565. |
| [陈正仁, 蓝开敏 (2003). 贵州杜鹃花属(杜鹃花科)一新种——荔波杜鹃. 植物分类学报, 41, 563-565.] | |
| [8] |
Doyle JJ, Doyle JL (1990). Isolation of plant DNA from fresh tissue. Focus, 12, 13-15.
DOI URL |
| [9] |
Earl DA, vonHoldt BM (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4, 359-361.
DOI URL |
| [10] |
Evanno G, Regnaut S, Goudet J (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14, 2611-2620.
DOI PMID |
| [11] | Fei SP, Zhou W, Chen ZR, Huang CL (2023). Effects of different hormone treatments on seed germination of Rhododendron liboense, a critically endangered plant. Guizhou Forestry Science and Technology, 51(2), 15-18. |
| [费仕鹏, 周玮, 陈正仁, 黄承玲 (2023). 不同激素处理对荔波杜鹃种子萌发的影响. 贵州林业科技, 51(2), 15-18.] | |
| [12] |
Frankham R (2003). Genetics and conservation biology. Comptes Rendus Biologies, 326, 22-29.
DOI URL |
| [13] | Gibbs D, Chamberlain DF, Argent G (2011). The Red List of Rhododendrons. Botanic Gardens Conservation Internationa, Richmond, UK. 56. |
| [14] |
Hewitt G (2000). The genetic legacy of the Quaternary ice ages. Nature, 405, 907-913.
DOI |
| [15] |
Holsinger KE, Weir BS (2009). Genetics in geographically structured populations: defining, estimating and interpreting FST. Nature Reviews Genetics, 10, 639-650.
DOI PMID |
| [16] |
Hu W, Zhang ZY, Chen LD, Peng YS, Wang X (2020). Changes in potential geographical distribution of Tsoongiodendron odorum since the Last Glacial Maximum. Chinese Journal of Plant Ecology, 44, 44-55.
DOI |
|
[胡菀, 张志勇, 陈陆丹, 彭焱松, 汪旭 (2020). 末次盛冰期以来观光木的潜在地理分布变迁. 植物生态学报, 44, 44-55.]
DOI |
|
| [17] |
Huang CL, Su CH, Tian XL, Chen ZR, Wen XY (2018). Discovery of two new populations of the rare endemic Rhododendron liboense in Guizhou, China. Oryx, 52, 610-611.
DOI URL |
| [18] |
Konar A, Choudhury O, Bullis R, Fiedler L, Kruser JM, Stephens MT, Gailing O, Schlarbaum S, Coggeshall MV, Staton ME, Carlson JE, Emrich S, Romero-Severson J (2017). High-quality genetic mapping with ddRADseq in the non-model tree Quercus rubra. BMC Genomics, 18, 417.
DOI PMID |
| [19] |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35, 1547-1549.
DOI PMID |
| [20] |
Li A, Ge S (2002). Advances in plant conservation genetics. Biodiversity Science, 10, 61-71.
DOI |
|
[李昂, 葛颂 (2002). 植物保护遗传学研究进展. 生物多样性, 10, 61-71.]
DOI |
|
| [21] | Lischer HEL, Excoffier L (2012). PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics, 2, 298-299. |
| [22] | Liu DT, Zhang L, Wang JH, Ma YP (2020). Conservation genomics of a threatened Rhododendron: contrasting patterns of population structure revealed from neutral and selected SNPs. Frontiers in Genetics, 11, 757. DOI: 10.3389/fgene.2020.00757. |
| [23] |
Liu XM, Fu YX (2015). Exploring population size changes using SNP frequency spectra. Nature Genetics, 47, 555-559.
DOI PMID |
| [24] | Liu YH, Wang HC, Yang J, Dao ZL, Sun WB (2024). Conservation genetics and potential geographic distribution modeling of Corybas taliensis, a small ‘sky Island’ orchid species in China. BMC Plant Biology, 24, 11. DOI: 10.1186/s12870-023-04693-y. |
| [25] | Ministry of Ecology and Environment of the People’s Republic of China, Chinese Academy of Sciences (2023). Comparison of China Biodiversity Red List—Higher Plants Volume (2020). [2023-5-18]. https://www.mee.gov.cn/xxgk2018/xxgk/xxgk01/202305/t20230522_1030745.html. |
| [中华人民共和国生态环境部, 中国科学院 (2023). 中国生物多样性红色名录——高等植物卷(2020). [2023-5-18]. https://www.mee.gov.cn/xxgk2018/xxgk/xxgk01/202305/t20230522_1030745.html.] | |
| [26] | Nagano Y, Tashiro H, Nishi S, Hiehata N, Nagano AJ, Fukuda S (2022). Genetic diversity of loquat (Eriobotrya japonica) revealed using RAD-Seq SNP markers. Scientific Reports, 12, 10200. DOI: 10.1038/s41598-022-14358-9. |
| [27] |
Ni J, Yu G, Harrison SP, Prentice IC (2010). Palaeovegetation in China during the late Quaternary: biome reconstructions based on a global scheme of plant functional types. Palaeogeography, Palaeoclimatology, Palaeoecology, 289, 44-61.
DOI URL |
| [28] |
Nielsen R (2000). Estimation of population parameters and recombination rates from single nucleotide polymorphisms. Genetics, 154, 931-942.
DOI PMID |
| [29] | Pan WT, Sun JJ, Yuan QQ, Zhang LL, Deng KQ, Li YQ (2022). Analysis of genetic diversity and structure in different provenances of Liriodendron by RAD-seq technique. Scientia Silvae Sinicae, 58(4), 74-81. |
| [潘文婷, 孙建军, 原勤勤, 张利利, 邓康桥, 厉月桥 (2022). RAD-seq技术研究鹅掌楸属种源遗传多样性和遗传结构. 林业科学, 58(4), 74-81.] | |
| [30] | Premarathne MDGP, Fukutome N, Yamasaki K, Hayakawa F, Nagano AJ, Mizuno H, Ibaragi N, Nagano Y (2021). Elucidation of Japanese pepper (Zanthoxylum piperitum De Candolle) domestication using RAD-Seq. Scientific Reports, 11, 6464. DOI: 10.1038/s41598-021-85909-9. |
| [31] |
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945-959.
DOI PMID |
| [32] |
Rochette NC, Rivera-Colón AG, Catchen JM (2019). Stacks 2: analytical methods for paired-end sequencing improve RADseq-based population genomics. Molecular Ecology, 28, 4737-4754.
DOI PMID |
| [33] |
Shen YT, Yao G, Li YF, Tian XL, Li SM, Wang N, Zhang CJ, Wang F, Ma YP (2024). RAD-seq data reveals robust phylogeny and morphological evolutionary history of Rhododendron. Horticultural Plant Journal, 10, 866-878.
DOI URL |
| [34] | Sun WB, Yang J, Dao ZL (2019). Study Conservation of Plant Species with Extremely Small Populations (PSESP) in Yunnan Province, China. Science Press, Beijing. 2-27. |
| [孙卫邦, 杨静, 刀志灵 (2019). 云南省极小种群野生植物研究与保护. 科学出版社, 北京. 2-27.] | |
| [35] | Wright S (1968). Evolution and the Genetics of Populations. University of Chicago Press, Chicago, USA. |
| [36] | Yang CH, Yao BF, Dai J (2019). Preliminary Investigation on Rhododendron liboense in Guizhou. Guizhou Forestry Science and Technology, 47(3), 14-17. |
| [杨成华, 姚炳矾, 代杰 (2019). 贵州荔波杜鹃初步调查. 贵州林业科技, 47(3), 14-17.] | |
| [37] |
Yoichi W, Tamaki I, Sakaguchi S, Song JS, Yamamoto SI, Tomaru N (2016). Population demographic history of a temperate shrub, Rhododendron weyrichii (Ericaceae), on continental islands of Japan and South Korea. Ecology and Evolution, 6, 8800-8810.
DOI URL |
| [38] | Zhang X, Zhang XJ, Ma YP, Li ZH, Wan YM, Ma H (2021). Genetic diversity assessment of Rhododendron sinofalconeri with genotyping by sequencing (GBS). Bulletin of Botanical Research, 41, 429-435. |
|
[张序, 张秀姣, 马永鹏, 李正红, 万友名, 马宏 (2021). 基于GBS简化基因组技术的宽杯杜鹃遗传多样性分析. 植物研究, 41, 429-435.]
DOI |
|
| [39] |
Zhang XJ, Liu XF, Liu DT, Cao YR, Li ZH, Ma YP, Ma H (2021). Genetic diversity and structure of Rhododendron meddianum, a plant species with extremely small populations. Plant Diversity, 43, 472-479.
DOI URL |
| [40] | Zhu XL, Tang JM, Jiang HD, Yang YS, Chen ZY, Zou R, Xu AZ, Luo YJ, Deng ZH, Wei X, Chai SF (2023). Genomic evidence reveals high genetic diversity in a narrowly distributed species and natural hybridization risk with a widespread species in the genus Geodorum. BMC Plant Biology, 23, 317. DOI: 10.1186/s12870-023-04285-w. |
| [1] | 殷斯, 杨依婷, 卢瑞玲, 念蕊, 郝转, 高永. 滇魔芋中国种群的谱系地理研究[J]. 植物生态学报, 2025, 49(2): 308-319. |
| [2] | 张望, 谭思仪, 涂文琴, 娄安如. 新疆吐哈地区梭梭种群遗传多样性和遗传结构[J]. 植物生态学报, 2025, 49(11): 1858-1868. |
| [3] | 王雨婷, 刘旭婧, 唐驰飞, 陈玮钰, 王美娟, 向松竹, 刘梅, 杨林森, 傅强, 晏召贵, 孟红杰. 神农架极小种群植物庙台槭群落特征及种群动态[J]. 植物生态学报, 2024, 48(1): 80-91. |
| [4] | 张宏祥, 闻志彬, 王茜. 新疆野苹果种群遗传结构及其环境适应性[J]. 植物生态学报, 2022, 46(9): 1098-1108. |
| [5] | 陈天翌, 娄安如. 青藏高原东侧白桦种群的遗传多样性与遗传结构[J]. 植物生态学报, 2022, 46(5): 561-568. |
| [6] | 王春成, 马松梅, 张丹, 王绍明. 柴达木野生黑果枸杞的空间遗传结构[J]. 植物生态学报, 2020, 44(6): 661-668. |
| [7] | 张新新, 王茜, 胡颖, 周玮, 陈晓阳, 胡新生. 植物边缘种群遗传多样性研究进展[J]. 植物生态学报, 2019, 43(5): 383-395. |
| [8] | 张俪文, 韩广轩. 植物遗传多样性与生态系统功能关系的研究进展[J]. 植物生态学报, 2018, 42(10): 977-989. |
| [9] | 王锦楠, 陈进福, 陈武生, 周新洋, 许东, 李际红, 亓晓. 柴达木地区野生黑果枸杞种群遗传多样性的AFLP分析[J]. 植物生态学报, 2015, 39(10): 1003-1011. |
| [10] | 刘军, 姜景民, 邹军, 徐金良, 沈汉, 刁松峰. 中国特有濒危树种毛红椿核心和边缘居群的遗传多样性[J]. 植物生态学报, 2013, 37(1): 52-60. |
| [11] | 张利锐, 彭艳玲, 任广朋, 周永锋, 李忠虎, 刘建全. 马尾松和黄山松两个核基因位点的群体遗传多样性和种间分化[J]. 植物生态学报, 2011, 35(5): 531-538. |
| [12] | 张炜, 罗建勋, 辜云杰, 胡庭兴. 西南地区麻疯树天然种群遗传多样性的等位酶变异[J]. 植物生态学报, 2011, 35(3): 330-336. |
| [13] | 魏源, 王世杰, 刘秀明, 黄天志. 不同喀斯特小生境中土壤丛枝菌根真菌的遗传多样性[J]. 植物生态学报, 2011, 35(10): 1083-1090. |
| [14] | 张云红, 侯艳, 娄安如. 华北地区小丛红景天种群的AFLP遗传多样性[J]. 植物生态学报, 2010, 34(9): 1084-1094. |
| [15] | 刘伟, 王曦, 干友民, 黄林凯, 谢文刚, 苗佳敏. 高山嵩草种群在放牧干扰下遗传多样性的变化[J]. 植物生态学报, 2009, 33(5): 966-973. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
Copyright © 2026 版权所有 《植物生态学报》编辑部
地址: 北京香山南辛村20号, 邮编: 100093
Tel.: 010-62836134, 62836138; Fax: 010-82599431; E-mail: apes@ibcas.ac.cn, cjpe@ibcas.ac.cn
备案号: 京ICP备16067583号-19
![]()